ISSN: 0973-7510
E-ISSN: 2581-690X
Recently, applying industrial bio-processing strategies are being a promised tool that used to produce large quantities of bioactive metabolites from different microbial cells. So in this study, the statistical experimental designs (Plackett–Burman and Box–Behnken) were used for optimization of local S. violaceoruber SYA3 biomass production (18g/l) and fabrication the highest nano-ZnO mass weight (15g/l). Since the nano-ZnO was fabricated by using the extracted periplasmic metabolites as a reducing agent and Zn(NO3)2.6 H2O as a precursor. The morphological properties and size of the nano-ZnO were evaluated using SEM and TEM analysis. Large-scale production was studied via batch fermentation mode using semi-industrial scale bioreactor (8.9L). The biomass production and the fabricated nano-ZnO was (22.8g/l) and (56.8g/l), respectively. Finally, this work reported developing a new cheap and eco-friendly nano-ZnO fabrication method and applying nano-ZnO as a strong anti-phytopathogens agent.
Biosynthesis of nano-ZnO, Plackett–Burman design, Box–Behnken design, Batch mode strategy, Anti-phytopathogens efficiency
Nanotechnology is the most innovative field of some industrial areas and now there are some nano-products was commercialized throughout the world1. Nanoparticles fabrication is carried out by physical, chemical and biogenic methods2. Recently, scientists found that the industrial production of some metal nanoparticles by applying physical/mechanical or chemical strategy may lead to the formation of the unwanted chemical moieties which may classified as a pollutant to our environment. So there are huge efforts to develop an eco-friendly nanoparticles fabrication method to reduce the production line cost, increase the final yield and improve its efficiency3,4,5,6. Recently, bio-fabrication of nanoparticles by using microbial cells considered as an environment-friendly, cost-effective and applicable method. Since microbial cells secreted some carbohydrates, amino acids and enzymes that supposed to act as reducing, capping and stabilization agents that used in bio-fabrication reaction7. The colloidal nano-ZnO solution was described as a bio-safe white solution which has a broad range of applications in several sectors such as agricultural sectors8. The high production cost of bioactive metabolites considered as the major bottlenecks facing industrial producers due to the cost of the microbial cultivation media3. So recently, there are more efforts to isolate the most potent microbe that produce promising bioactive compounds and to optimize its culturing medium to reduce the production costs drastically3,6. There are few publications that studied the Streptomyces sp as an efficient candidates that used for the bio-fabrication of nano-ZnO by using intracellular or extracellular bioactive metabolites fractions as reducing and stabilizing agents9. Statistical-mathematical designs such as Plackett-Burman and Box-Behnken designs reported recently as a more balanced alternative strategy to the one-factor-at-a-time approach5. These statistical-mathematical designs depended on specific sequence steps such as screening of the most effective factors, studying their interaction effect and calculating their optimum concentrations that produce the highest quantities and/or qualities of the desired response(s). In an industrial bioprocesses strategy, living microbial cells used as cell factories that produce bioactive metabolites by utilizing mathematical kinetic models which are able to develop these cell properties and/or the final production bioprocesses4,9. Kinetic models used to understand, predict, develop and assess the effects of modifying (adding and/or removing) cell cultivation components and/or cultivation conditions4,5. For an agricultural application, the current investigations focused on using different statistical bioprocess strategies for cultivation of local Streptomyces sp to get the most effective metabolites (reducing and stabilizing agent) to develop the more safely biochemical synthesis pathway for nano-ZnO fabrication then applied nano-ZnO as an anti-phytopathogens agent. Also, this work includes, scaling up strategy performed using semi-uncontrolled batch (shake-flask) and controlled batch systems.
Collection of soil samples and isolation of Actinomycetes
Soil samples were collected from different Egyptian cultivated fields (Alexandria, Dakahlia, Sharkia and Menofya governorates). By using serial dilution method and starch nitrate agar the Actinomycetes were isolated and purified. Starch nitrate agar medium composed of (g/l): Starch, 20; KNO3, 2; K2HPO4, 1; MgSO4.7H2O, 0.5; NaCl, 0.5; CaCO3, 3; FeSO4.7H2O, 0.01; and agar, 20. The purified colonies conserved in slants at 4°C for further work10,11.
Nano-ZnO biofabrication reaction and its anti-phytopathogenic activities
The nano-ZnO biofabrication reaction was prepared by using the microbial bioactive metabolites that extracted from different isolates. In a 250 ml conical flask, 10 m1 of Zn(NO3)2•6H2O solution (1M) was added to 90 ml of the extracted metabolites and left until the colloidal white solution was formed. The settled down white precipitate was washed with distilled water for 2–3 times and ethanol for 3–4 times. The oven dried nano-ZnO (50°C) was scraped out and prepared for studying its antimicrobial activity by using well diffusion method against some phytopathogenic bacteria (Erwinia amylovora, Acetobacter pasteurianus) and fungi (Aspergillus flavus, Phytophthora arenaria). Fifty µl of nano-ZnO (1M) was placed into wells and after full diffusion; the plates were spread with different pathogens. The plates were incubated at 30-37°C for 24-96 hr and examined for zones of inhibition3,4,5. Triplicates were maintained and the average values were calculated to determine the ultimate antimicrobial activities. Finally, the ANOVA analysis was used to demonstrate a significant difference (p< 0.05).
Characterization of bio-fabricated nano-ZnO
The UV-Vis absorption spectra was recorded using (Biochrom Libra S50, UV/V is Spectrophotometer, England) spectrophotometer which equipped with “UVWinlab” software to record data and the numerical data were plotted by using Sigma plot 12.5. The structural properties (real estate properties) and size of the nano-ZnO were analyzed using scanning electron microscope (Jeol JSM-6360LA) and transmission electron microscope (JEOL JEM-2100, Japan).
Molecular identification of the selective Actinomycetes isolate
The isolate which produced the proficient nano-ZnO that showed a strong antimicrobial efficiency was selected and subjected to further observations. The genomic DNA was isolated by a modified method of12,13,14. The PCR reaction conditions were performed according to10,15,16 by using 35 cycles of 94°C (1min), 55°C (1min) and 72°C (2min) and then a 7min termination at 72°C. PCR products were purified using the PCR purification kit (Qiagen, Germany) and sequenced by MacroGen Co. (Seoul, Korea). Furthermore, the determined sequences were compared with the sequences deposited in the NCBI Gene Bank by using BLAST software. Multiple sequence alignment and the phylogenetic tree were analyzed and illustrated by using BioEdit software (2006) and the MEGA 6.0 program (2013).
Screening of the most effective microbial fraction for bio-fabrication of nano-ZnO
The localization of the effective microbial metabolites (reducing/stabilizing agent) that extracted to fabricate the highest nano-ZnO mass weight was determined by using different cellular compartment extracts (extracellular, intracellular or periplasmic) according to15,17,18. The modified Bennett’s Broth medium (ISP2) that consisted of (g/l): yeast extracts 4; malt extract 10 and dextrose 10 (pH 7.3) was inoculated with the identified Streptomyces strain and incubated at 30°C, under shacking conditions (200 rpm). The grown culture was filtered using the suction filtration system to prepare the extracellular fraction and the residual cells used for preparing the periplasmic and intracellular fractions. To obtain the periplasmic fraction, the isolate pellet washed extensively using Milli-Q deionized water to remove any medium component and debris then the washed pellet was added to 100 ml distilled water in an Erlenmeyer flask and agitated at 200 rpm for 48 hr at 30 °C. The periplasmic fraction was obtained by centrifugation at 10,000 rpm for 15 min at 4°C, then the periplasmic fraction (supernatant) was collected and kept at 4°C for further work4.
Optimization of biomass production by using statistical experimental designs
From the previous publications, the four formulated media were chosen to cultivate the identified strain and select the more proficient medium that produces the largest cell density and the maximum metabolites yield to fabricate the highest nano-ZnO mass weight19,20,21,22. Medium No. 1 (ISP1) consists of (g/l): dextrose 10, yeast extract 3, and tryptone 5, (pH 7.0), Medium No.2 (ISP2) consists of (g/l): yeast extract4; malt extract 10, peptone 5 and dextrose 10 (pH 7.3), Medium No.3 consists of (g/l): glucose 10, yeast extract 20, peptone 5, KH2PO4 2, K2HPO4 4, and MgSO4.7H2O 0.5, (pH 6.8), and finally Medium No.4 consists of (g/l): Dextrose 10; malt-extract 3; yeast-extract 3; peptone 5 (pH 7.0). All of these media sterilized for 15 min at 121ºC, inculated with the identified strain (initial O.D600 was ~ 0.3) and finally the flasks incubated at 30°C, under shacking conditions (200rpm). The growth curve was monitored and the dry cell mass weight (DCMW) was determined.
Plackett-Burman design
In this study, Plackett-Burman design was used to select the factors that influence on the DCMW production significantly in order to obtain the more manageable set of variables that represented at two levels (+1, and -1). All trials were set up in 250 ml Erlenmeyer flasks containing 50 ml of the chosen medium. The significant variables were determined by calculating the p-value (p<0.05) through standard regression analysis. Also, the estimated main effect was used to elucidate the significance of variables as described previously4,5 by using the 1st order model equation (Eq. 1). The predicted optimum levels of the independent variables were validated and compared to the basal condition.
![]()
…(1)
Where: Y is the response (DCMW), bo is the model intercept, bi is the estimated variable; and Fi represents the factor.
Response surface methodology
In this work, Box-Behnken modeling and its analysis were carried out to study the interactions among the significant variables that represented at three levels (+1, 0 and -1). The fifteen combinations were performed and the average of DCMW (g/l) was calculated for each trial and the coefficient results of each variable were determined by using the second-order polynomial structured equation (Eq. 2).The simultaneous effects of the most significant independent factors on each response were visualized using three-dimensional graphs generated by using Sigma plot 12.5 software. The value of each X1, X2 and X3 were further optimized to calculate the best Y value using Microsoft excel 2007 solver. All experiments were carried out in triplicates and the average was recorded. To prove the accuracy of the model, the % accuracy was calculated by using the Eq. (3).
Y = b0 + b1X1 + b2X2 + b3X3 + b11X12 + b22X22 + b33X33+ b12X1 X2 + b13 X1 X3 + b23X2X3
…(2)
Where: Y is the response (DCMW); bo is the model intercept; X1, X2 and X3 are independent variables; b1, b2 and b3 are linear coefficients; b12, b13 and b23 are cross product coefficients; and b11, b22 and b33 are the quadratic coefficients.
Accuracy of the model = [YExperiment/ YCalculated] x 100
…(3)
Large-scale fermentation experiments
The behavior of microbial cell can be described kinetically according to the type of cultivation mode (batch and fed-batch cultivation). Therefore, the growth kinetic relationship is affected by many parameters such as a composition of media, substrate concentration, specific growth rate and feeding strategy into a bioreactor5. In this work, scaling up strategy was made by using semi-uncontrolled batch (shake-flask) and controlled batch cultivation.
Cultivation in shake-flask
The standard procedure for batch fermentation mode in shake flask was conducted by inoculating the flask (500 ml) that contained the optimized broth medium (250 ml) with a pure culture of the tested cells, and the initial optical density (600 nm) was fixed at ~ 0.3 then this flask incubated at 30°C under shaking condition (200rpm). The samples were taken periodically and the dry cell mass was prepared by using centrifugation at 10,000 rpm for 20 min, then the pellet washed twice with 0.9% (w/v) and dried at 105°C until getting the constant dry cell mass weight.
Batch cultivation strategy
In this investigation, batch fermentation mode was conducted in a 8.9 L bench-top with maximum 7 L working volume bioreactor (Winpact Bench-Top Fermentor, FS-01-B Series, Version: V2.4, Cleaver Scientific Ltd, U.S.A.). The bioreactor vessel containing 4600 ml of the statistically optimized medium that sterilized at 121°C for 25 min, while the glucose solution was sterilized separately at 121°C for 15min then added aseptically into the sterilized bioreactor. The fermentation temperature was adjusted at 30°C and the agitation speed & aeration rate started at 150 rpm & 1.0 vvm respectively and the dissolved oxygen concentration was maintained above 40% of air saturation. The pH was controlled at 6.8 by adding NaOH (5 N) or HCl (5 N). Antifoam (silicone oil 0.5:10 v/v) was added after 7hr of the inoculation. Yield coefficient (YX/S) was determined depending on the biomass yield and amount of the consumed glucose by using Eq.(4). In the log phase, cell mass density increases exponentially with time (t), so the specific growth rate μ (h-1) was calculated by using Eq.(5).
![]()
…(4)
![]()
…(5)
Bioapplication of fabricated nano-ZnO as Anti phytopathogens agent In vitro
The fabricated nano-ZnO solution (50 µl of 1 M) was applied as antimicrobial agent by using well diffusion method against phytopathogenic bacteria (Erwinia persicina and Erwinia carotovora) and phytopathogenic fungi (Fusarium moniliforme and Helminthosporium sp). An ANOVA demonstrated a significant difference (p< 0.05) for all the obtained results.
In recent years, nanobiotechnology is an escalating field of the modern research involving nanoparticles fabrication strategy, characterization, large-scale production, and its application by controlling the nano-shape and/or the nano-size3,4,5,23. Several complex sets of engineering and scientific challenges in the bioprocessing industries for safe manufacturing the highest quality of non-particles by using the proficient in expensive methods can be solved through nano-biotechnological strategies. These challenges can be addressed, by producing the most effective bioactive compounds and optimizing the fabrication reaction, through applying new industrial biotechnological strategies3,4. Cost-effective bioprocessing strategies for the biosynthesis of nano-ZnO by using the extracted microbial metabolites were used in this work to produce the new simple and relatively inexpensive anti-phytopathogens agent.
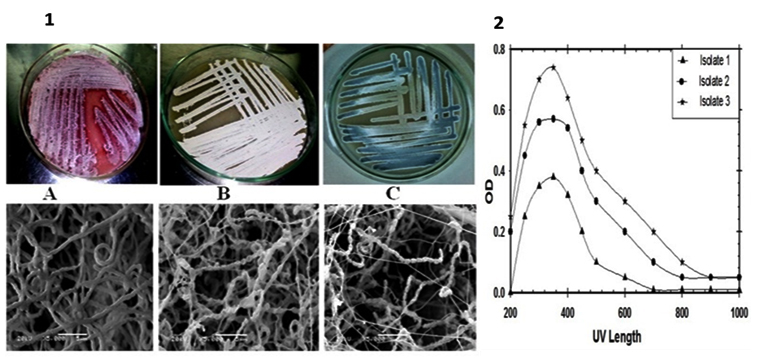
Fig. 1. Digital photographs indicates color of the aerial mycelium of the selected isolates that grown on starch nitrate medium for 7–14 days at 30°C and scanning electron micrograph showing their spore-chain morphology and their spore-surface ornamentation at constant magnification; (A): isolate1, (B): isolate2 and (C): isolate3. (2)UV-visible spectroscopyanalysis for different biofabricated nano-ZnO.
Screening the efficiency of bio-fabricated nano-ZnO
In this study, a total of fifty-five actinomycetes isolates were screened for biosynthesis of nano-ZnO by using extracted bioactive compounds as reducing agent and zinc nitrate hexahydrate as a precursor. Of them, three different isolates produced nano-ZnO as shown in Fig.(1). According to Kakhaki et al., and Gupta et al.,24,25, the nano-ZnO absorption spectra determined at 300-400 nm depending on the size of Zn nanoparticles. So in this investigation UV-visible absorption spectroscopy technique used to examine and confirm the optical properties of bio-fabricated nano-ZnO. The strong absorption spectra of all bio-fabricated nano-ZnO were shown at ~ 360 nm. The nano-ZnO antimicrobial activities were determined by calculating the inhibition zones as shown in Table (1). Generally, nano-ZnO that fabricated by using isolate No 3 extract showed more effective antimicrobial activity against all tested phytopathogens (P< 0.05). So this isolate was selected for further investigations. First of all, this isolate was identified by using PCR-sequencing technique and the sequence of nucleotide was deposited into the database of GenBank as Streptomyces violaceoruber SYA3 (http://www.ncbi.nlm.nih.gov/nuccore/1036392258). A phylogenetic tree based on 16S rDNA gene sequences of members of the genus Streptomyces was constructed according to the neighbor-joining method of Saitou and Nei26 with the software package MEGA 6.0 as shown in Fig. (2).
Fig. 2. Phylogenetic tree constructed using 16S rDNA gene sequence analysis by using the neighbor-joining method with the aid of MEGA 6.0 program, showing the relationship of ▲ Streptomyces violaceoruber strain SYA3 with reference strains
Table (1):
Antagonistic effect of nano-ZnO fabricated by using different isolates against some phytopathogens.
| Type | Pathogens | Zone of inhibition (mm) | ||
|---|---|---|---|---|
| Isolate 1 | Isolate 2 | Isolate 3 | ||
| Phytopathogenic bacteria | Erwinia amylovora | 23.0±0.3 | 10.0±0.3 | 25.0±0.9 |
| Acetobacter pasteurianus | 10.0±0.3 | 20.0±0.5 | 30.0±0.9 | |
| Phytopathogenic fungi | Aspergillus flavus | nil | nil | 30.0±0. 5 |
| Phytophthora arenaria | 18.0±0.4 | 5.0±0. 5 | 31.0±2.3 | |
The data represents mean values of three independent experiments± SD
Characterization of nano-ZnO biofabricated from S.violaceoruber strain SYA3
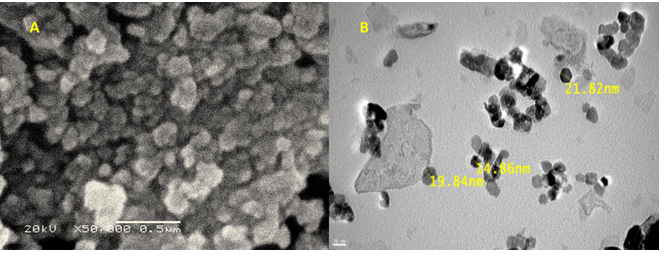
In this work, the surface morphology of nano-ZnO (shape and size) was clearly characterized by using SEM and TEM analysis as shown in Fig. (3). The shape of nano-ZnO was asquasi-spherical particles as shown by SEM analysis Fig. (3A), with a narrow size distribution and the average size was calculated as 18.8 nm as indicated by Fig. (3B). These results better than the final reports6,27,28 which fabricated the crystalline spherical zinc nanoparticles from fungi with size ranged from 54.8- 82.6nm.
Fig. 3. SEM (A)and TEM (B) analysis for the fabricated nano-ZnO. Scale bar 0.5μm and 20nm respectively
Cellular compartmentalization of the most potent bioactive metabolite extracts
The microbial cells are able to produce primary and secondary metabolites 4,30. Generally, metabolites may be exported (leakage) from the microbial cells directed to the medium or periplasmic space or still found in the cytoplasm of the cells; because their translocation depends on the nature domain of the target bioactive compounds31,32,33. To detect the occurrence of the proficient bioactive compounds that used as reducing and stabilizing agents in the fabrication of nano-ZnO; the extracellular, periplasmic, or intracellular small-scale fractions were tested and the average dry weight of the fabricated nano-ZnO was determined. The final results indicated that the periplasmic fraction was given the highest nano-ZnO quantity.
Bioprocessing strategies for biomass production of S.violaceoruber strain SYA3
As a general, the success of industrial production for biological control agents depending not only on the microbial cells, characterization and its pathogenicity but also on the successful mass production of the microbial cells. The selection of a best carbon source that able to enhance microbial growth without leading to catabolism is an important step in biomass optimization strategy. The relation between bioactive compound, protein secretion and the growth profile of the microbial cells is a key consideration for increased nanoparticles production. Hence, the growth medium could be one of the main factors affecting cells/protein production during a fermentation process31,34. Therefore, the use of high cell density cultivation is required in order to improve nano-ZnO biosynthesis. Using the modern biotechnological strategies to get the most suitable and the cheapest S. violaceoruber strain SYA3 cultivation medium to fabricate the largest nano-ZnO mass weight was the main target of all the following experiments. Firstly; the growth curve of S.violaceoruber strain SYA3 that cultivated in different industrial media was investigated. The final results indicated that the medium No. 3 was the best cultivated medium that produce the largest bioactive metabolites that used for fabrication of the highest nano-ZnO mass weight. The final results indicated that there is a direct correlation between DCM and nano-ZnO productions, so thereafter other experimental studies that performed in this work, focused on increasing DCM production by using bioprocessing strategies (different statistical experimental designs). Nowadays application of the statistical experimental designs has become a very attractive, effective tool for optimization of the bioactive metabolite production that reported in many articles3,5. The statistical optimization strategies for biomass production were carried out in two steps by using a fractional factorial method (Plackett–Burman design) followed by response surface method (Box–Behnken experimental design). To the best of our knowledge, there has been no work concerning the evaluation of such a strategy for improving nano-ZnO production which bio-fabricated by using S.violaceoruber strain SYA3 extracts.
Table (2):
Randomized Plackett-Burman experimental design matrix used for evaluation of six independent variables along with the S.violaceoruber strain SYA3 biomass production.
| Run | Variables (g/l) | Biomass dry weight (g/l) | |||||
|---|---|---|---|---|---|---|---|
| Glucose | Peptone | Yeast extract | MgSO4.7H2O | KH2PO4 | K2HPO4 | ||
| 1 | 1(20) | 1(6) | -1(2) | -1(0.1) | -1(1) | 1(6) | 3.9 |
| 2 | 1(20) | -1(3) | -1(5) | -1(0.1) | 1(4) | 1(6) | 4.0 |
| 3 | -1(10) | -1(3) | -1(2) | 1(2) | 1(4) | 1(6) | 3.9 |
| 4 | -1(10) | -1(3) | 1(5) | 1(2) | 1(4) | 1(6) | 4.1 |
| 5 | -1(10) | 1(6) | 1(5) | 1(2) | 1(4) | -1(2) | 4.2 |
| 6 | 1(20) | 1(6) | 1(5) | 1(2) | -1(1) | -1(2) | 4.2 |
| 7 | 1(20) | 1(6) | 1(5) | -1(0.1) | -1(1) | -1(2) | 4.1 |
| 8 | -1(10) | -1(3) | -1(2) | -1(0.1) | -1(1) | -1(2) | 3.8 |
The Plackett-Burman design was adopted for screening the most significant nutritional parameters for biomass and bioactive metabolites production by using S.violaceoruber strain SYA3 in submerged fermentation. In the experimental design, six independent variables were screened to pick significant factors that influence biomass production by representing them at two levels (high and low) in eight trial experiment. Table (2) showed the randomized Plackett-Burman design matrix and the final DCM production as a response which revealed a wide variation. Based upon the regression coefficient analysis of testing variables glucose (F1), peptone (F2), yeast ext. (F3), MgSO4.7H2O (F4) and KH2PO4 (F5) were shown the positive effect on DCM production, where K2HPO4 (F6) was contributed negatively. For each variable, the responses mean of the experiments with high value (+1) has been calculated using the formula ∑ (+1)/n (+1), while the responses mean of the experiments with the low value (-1) have been calculated using the formula ∑ (-1)/n (-1). The main effects of the examined factors and the DCM were calculated from Eq.(6). According to the estimated main effect, the six variables were analyzed using linear multiple regression analysis methods. The P-value was calculated to determine the significant variables and the final results were summarized in Table (3). The multiple linear regression models of Plackett-Burman screening method describes the relationship between the DCM production of S.violaceoruber strain SYA3 and six independent variables. The fitted model was generated as shown in Eq. (8). The final Plackett-Burman design step was the verification experiments, that examined by using the predicted levels of the independent variables and the final results compared to the basal condition. The final DCM production was (9.5 g/l) which is 2.8 times higher when eliminating peptone and K2HPO4 from the optimized medium, so the final medium composition consisted of (g/l) glucose 20.0g, yeast extract 5.0g, MgSO4.7H2O 2.0g and KH2PO4 4.0g. The significant independent variables glucose (X1), yeast extract (X2) and KH2PO4 (X3) were selected for further optimization method (Box-Behnken design) as shown in Table (4).
Table (3):
Statistical analysis of Plackett–Burman results.
| Variables | Coefficients | Standard Error | t Stat | P-value | |
|---|---|---|---|---|---|
| Glucose | 0.079167 | 0.002946 | 26.87005769 | 0.023681605* | |
| Peptone | 0.016667 | 0.002946 | 5.656854249 | 0.111388754 | |
| Yeast ext. | 0.102083 | 0.002946 | 34.64823228 | 0.018368703* | |
| MgSO4.7H2O | 0.052083 | 0.002946 | 17.67766953 | 0.035974313 | |
| KH2PO4 | 0.079167 | 0.002946 | 26.87005769 | 0.023681605* | |
| K2HPO4 | -0.03333 | 0.002946 | -11.3137085 | 0.056123917 | |
| df | SS | MS | F | Significance F | |
| Regression | 6 | 0.233021 | 0.038836806 | 1491.33 | 0.0198 |
| Residual | 1 | 2.6E-05 | 2.60417E-05 | ||
| Total | 7 | 0.233047 |
Table (4):
Box–Behnken experimental design matrix of three variables with actual factor levels corresponding to coded factor levels, and the mean of S.violaceoruber strain SYA3 biomass dry weight.
| Trial No. | Variables (g/l) | Biomass dry weight (g/l) | ||
|---|---|---|---|---|
| Glucose | Yeast extract | KH2PO4 | ||
| 1 | 0(20) | 0(5) | 0(4) | 9.3 |
| 2 | -1(15) | -1(3) | 0(4) | 8.6 |
| 3 | 0(20) | 1(15) | 1(10) | 10 |
| 4 | 0(20) | -1(3) | 1(10) | 7.2 |
| 5 | 0(20) | 0(5) | 0(4) | 9.3 |
| 6 | 1(30) | 1(15) | 0(4) | 12.8 |
| 7 | 1(30) | 0(5) | 1(10) | 8.6 |
| 8 | 1(30) | 0(5) | -1(2) | 10 |
| 9 | -1(15) | 1(15) | 0(4) | 13 |
| 10 | -1(15) | 0(5) | 1(10) | 8.7 |
| 11 | 0(20) | 1(15) | -1(2) | 14.3 |
| 12 | 0(20) | -1(3) | -1(2) | 9.3 |
| 13 | -1(15) | 0(5) | -1(2) | 11.3 |
| 14 | 0(20) | 0(5) | 0(4) | 9.3 |
| 15 | 1(30) | -1(3) | 0(4) | 8 |
Main effect = ∑ (+1)/n (+1) -∑ (-1)/n (-1)
…(6)
The confidence level%= 100*(1- P value)
…(7)
Y= 3.99+0.2 * Glucose + 0.01* Peptone + 0.09 * Yeast extract + 0.04 * MgSO4.7H2O + 0.07* KH2PO4 – 0.03 * K2HPO4
…(8)
The fifteen trials of Box-Behnken design experiment were analyzed using linear multiple regression analysis methods and the percentage of the confidence level (%) was calculated by using Eq. (7), summarized in Table (5). The final results showed that the higher levels of yeast extract and glucose support the highest DCM production. On the other hand, the lower levels of the DCM production were attained with increasing the KH2PO4 especially at lower levels of yeast extract and glucose. A second-order polynomial equation (Eq. 9) was fitted to predict the optimal point of the DCM production (the maximum point of the polynomial model) by using the Solver function in Microsoft Excel. The optimum levels of the three variables were estimated and found to be for the glucose; 27.5 g/l, yeast extract; 15 g/l, MgSO4.7H2O 2 g/l, and KH2PO4; 5 g/l that produced DCM 14.8 g/l which increased 4.5 times (model accuracy was 98%). After the culture medium ingredients improvement, the increase of the microbial biomass productivity can be achieved successively by optimizing the large scale fermentation conditions.
Table (5):
Statistical analysis of Box-Behnken results.
| Variables | Coefficients | Standard Error | t Stat | P-value | Confidences level% |
|---|---|---|---|---|---|
| Glucose (X1) | -0.275 | 0.158902 | -1.73062 | 0.144076 | 85.59 |
| Yeast ext. (X2) | 2.125 | 0.158902 | 13.37298 | 4.18E-05* | 99.9* |
| KH2PO4 (X3) | -1.3 | 0.158902 | -8.18112 | 0.000444* | 99.96* |
| X1X2 | 0.1 | 0.224722 | 0.444994 | 0.674927 | 32.51 |
| X1X3 | 0.3 | 0.224722 | 1.334982 | 0.239437 | 76.06 |
| X2X3 | -0.55 | 0.224722 | -2.44747 | 0.058117 | 94.19 |
| X1X1 | 0.375 | 0.233898 | 1.603262 | 0.16978 | 83.02 |
| X2X2 | 0.925 | 0.233898 | 3.954713 | 0.0108* | 98.92* |
| X3X3 | -0.025 | 0.233898 | -0.10688 | 0.919037 | 8.10 |
| df | SS | MS | F | Significance F | |
| Regression | 9 | 55.414 | 6.157111 | 30.48075 | 0.000763 |
| Residual | 5 | 1.01 | 0.202 | ||
| Total | 14 | 56.424 |
Y= 9.3 -0.3*X1 +2.2*X2 -1.3*X3 -0.05* X12 +0.43*X22 -0.63*X32 + 0.28*X1X2 +0.88* X1X3 -0.05* X2X3
…(9)
Large scale fermentation strategies
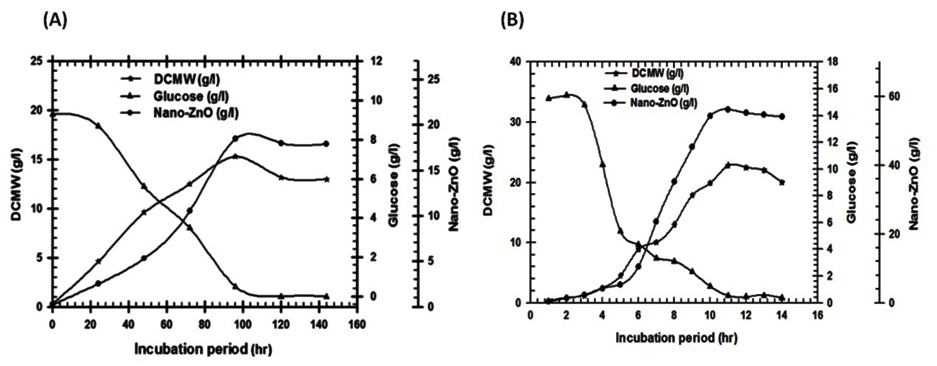
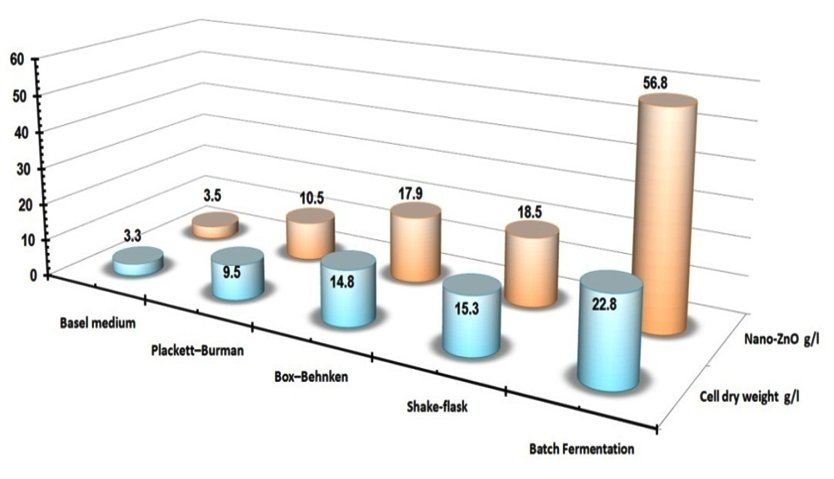
The most common fermentation system is the batch culture. This is a closed system in which there is no input or output of materials. The microbial population cell density increases constantly until exhaustion of some limiting factor, while other nutrient components of the culture medium decreased in time35,36. Successful scaling-up strategies are developed subsequently under small-scale batch cultivations, a pilot scale and then an industrial scale to increase the biomass productivity hence the extracted bioactive metabolites35,36. Since the fermentation parameters are studied initially by using small-scale batch cultivation (shake-flask) and then optimized through controlled large-scale fermentation modes 36,37,38. In this study, to scale up the cultures to higher cell densities, cultivation modes were performed: batch cultivation in a shake-flask, batch cultivation in 8.9 L bench-top bioreactor to estimate and comprises the growth kinetics at different submerged culturing systems as shown in Table (6). Shake-flask batch cultivation experiment was aimed to monitor the fabricated nano-ZnO and biomass profiles with consumed carbon source (glucose) which used to determine the correlations between the specific growth rate and the nano-ZnO fabrication rate Fig. (4A). By using kinetics measurements for S.violaceoruber strain SYA3 cells; the specific growth rate (µmax), the maximum biomass production (Xmax) and the maximum fabricated nano-ZnO (Pmax) were calculated and recorded as 0.142, 15.3 g/l, 18.5 g/l respectively at 144 hr. By using 8.9 L bench-top bioreactor the S.violaceoruber strain SYA3 biomass production was 22.8 g/l that extracted the highest bioactive metabolites that used as reductant for the fabrication of nan-ZnO (56.8 g/l). In this work, to scaling-up the tested strain to higher cell densities, cultivation strategy were improved to estimate the best growth kinetics by using this way: the statistical experimental designs were applied firstly then batch cultivation systems were optimized via a shake-flask, and 8.9 L bench-top as summarized in Fig.(5).
Fig. 4. (A) Real time monitoring for consumption of glucose, fabricated nano-ZnO and S.violaceoruber strain SYA3 growth pattern that cultivated in shake flask (500ml) by using submerged fermentation system. (B): Time course of batch mode for S.violaceoruber strain SYA3 that cultivated in Cleaver fermentor (8.9 L) by using submerged fermentation system
Fig. 5. Summary of the maximum DCM weight of S.violaceoruber strain SYA3 and the highest nano-ZnO mass weight that produced during statistical optimization process and large scale strategy
Table (6):
Comparison study between the cultivations of S.violaceoruber strain SYA3 in shake flask and batch fermentation mode in stirred tank bioreactor indicated by growth conditions, biomass and nano-ZnO production kinetics.
| Flask | Batch | |
|---|---|---|
| Container volume (ml) | 500 | 7000 |
| Working Volume (ml) | 250 | 3000 |
| Ph | Un-controlled | 6.8 |
| Aeration (vvm) | Un-controlled | Adjusted to let the dissolved O2 level did not below 40% |
| Agitation (RPM) | 200 | |
| Incubation period (hr) | 144 | 11 |
| Specific growth rate (µmax) | 0.142 | 0.096 |
| Biomass yield coefficient (YX/S) | 0.44 | 1.12 |
| Maximum biomass (Xmax) | 15.3 | 22.8 |
| Doubling time (td) | 25.45 | 4.234 |
| Maximum Product (Pmax) | 18.5 | 56.8 |
| Product yield coefficient (YP) | 0.3 | 3.75 |
Bioapplication of the fabricated nano-ZnO as antiphytopathogen agent in vitro
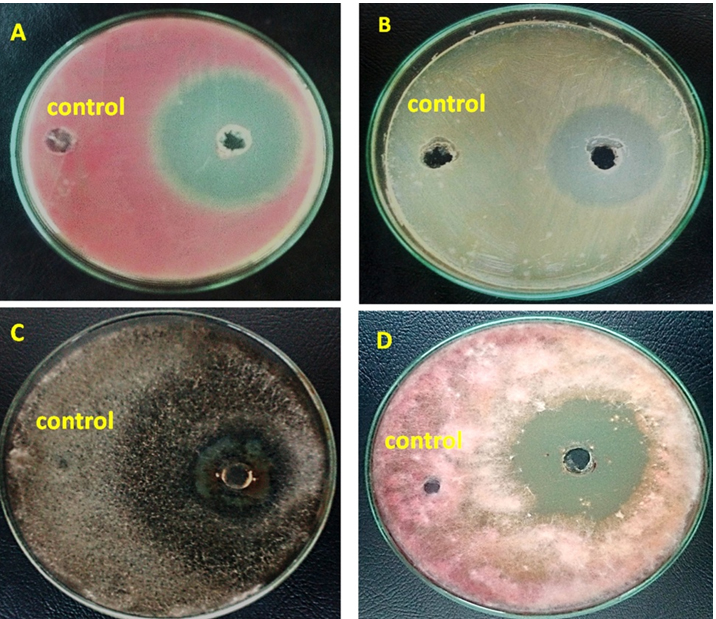
There are some phytopathogens difficult to control because they have developed resistance to many conventional pesticides. To overcome this resistance, it is important to explore novel anti-phytopathogens agents, which may replace or develop the current biocontrol strategies. Recently, for agricultural applications, nanomaterial studied as an effective nanobiofertilizers in seed germination as well as a promising biocontrol agent31,32. In this work the inhibitory effect of pure biofabricated nano-ZnO was evaluated by calculating the zone of inhibition that statistically analyzed. As a general, the results indicated that the fabricated nano-ZnO (50 µl of 1 M) showed proficient antimicrobial activities against all tested phytopathogens. The highest bacterial inhibitory effect was observed against Erwinia carotovora followed by Erwinia persicinaand where the highest fungal inhibitory activities were recorded against Fusarium moniliforme then Helminthosporium sp. Finally, these results revealed that the pure nano-ZnO showed the highest antibacterial activities (33-54mm) compared to the fungal inhibitory effect (20-40mm) as shown in Table (7) and Fig. (6). These recorded results more effective than that reported where the inhibition zones (14-32mm) was recorded by using hydrothermal synthesized nano-ZnO (30mg/ml) as antiphytopathogen agent28. Shah, Yehia, Wani, and He 2,8,33,39 reported that the nano-ZnO brought a significant inhibition in the spores germination of Alternaria alternata, Fusarium oxysporum, Rhizopus stolonifer, Mucor plumbeus and Penicilium expansum. Also, there are some studies reported the significant inhibition effect of ZnONPs (70nm) against postharvest pathogenic fungi (Botrytis cinerea and Penicillium expansum), and their mode of action was confirmed by SEM and Raman spectroscopy. Since the nano-ZnO caused deformation of fungal hyphae and prevent the development of the conidiophores and conidia which ultimately leads to the death of fungal cells29,33. Others suggested that the nano-ZnO may interact with microbial enzymes, proteins, or DNA to inhibit cell proliferation. Until now, the actual mechanism of action is not yet cleared 31,32.
Fig. 6. Antibacterial activities of the pure nano-ZnO against the growth of some phytopathogenic bacteria; (A) Erwinia carotovora and (B) Erwinia persicina; (Control) 0.1 mM of Zn (NO3)2·6H2O. Antifungal efficiency of pure nano-ZnO against the growth of some phytopathogenic fungi; (C) Fusarium moniliforme and (D) Helminthosporium sp.
Table (7):
Antimicrobial activity of nano-ZnO biofabricated by S.violaceoruber strain SYA3 against the growth of certain phytopathogen.
| Type | Patgogen | Inhibition zone (mm) |
|---|---|---|
| Phytopathogenic bacteria | Erwinia carotovora | 30.4±0.05 |
| Erwinia persicina | 50.0±0.06 | |
| Phytopathogenic fungi | Fusarium moniliforme | 40.6±0.05 |
| Helminthosporium sp | 20.5±0.21 |
The data represents mean values of three independent experiments± SD.
By a stepwise approach, the medium ingredients were evaluated by using statistical experimental designs (Plackett–Burman and Box–Behnken) to produce the highest S.violaceoruber strain SYA3 biomass production (14.8 g/l) and then fabricate the highest quantities of nano-ZnO (18.5 g/l) that used as anti-phytopathogens. Secondly, the scaling-up fermentation strategy was performed in a batch mode, then the maximum biomass production (Xmax) and nano-ZnO (Pmax) mass weight were recorded as 22.8 g/l, and 56.8 g/l respectively. Finally, the reported results open a new avenue for the biofabrication of nano-ZnO that used as antiphytopathogen agent by using Streptomyces spp., as a proficient and safe candidate.
ACKNOWLEDGMENTS
The authors acknowledge the Bioprocess Development Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg El-Arab, Alexandria, Egypt for funding the activities presented in this study. They also recognize the valuable technical help of the Central Laboratory, Advanced Technology and New Materials Research Institute, City for Scientific Research and Technological Applications, Alexandria, Egypt.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Muthu C R R, Rajalaxshmi A. Green Synthesis, Characterization of ZnO nanoparticles and Ceion doped ZnO nanoparticles assisted Sesbania grandiflora for photocatalytic application. Res J Material Sci, 2016; 4(2): 1-6.
- Shah M, Fawcett D, Sharma S, Tripathy K S, Poinern E J G. Green synthesis of metallic nanoparticles via Biological Entities. Materials, 2015; 8(11): 7278–7308.
- EL-Moslamy H S, Shokry H H, Rezk H A, Abdel-Fattah R Y. Bioprocess Strategies and Characterization of Anti- Multidrug Resistant Human Pathogens Copper/Copper Oxide Nanoparticles from Citrus Peel Waste Extracts. J Nanomater Mol Nanotechnol, 2017; 6: 1.
- EL-Moslamy H S, Elkady F M, Rezk H A, Abdel-Fattah R Y. Applying Taguchi design and large-scale strategy for mycosynthesis of nano-silver from endophytic Trichoderma harzianum SYA.F4 and its application against phytopathogens. Sci Rep, 2017; 7: 45297.
- EL-Moslamy H S, Kabeil S A S and Hafez E E. Bioprocess development for Chlorella vulgaris cultivation and biosynthesis of anti-phytopathogens silver nanoparticles. J Nanomater Mol Nanotechnol, 2016; 5: 1.
- Jeffina B J, Levin A G. Biosynthesis and Characterization of ZnO nanoparticle using Cassia auriculata Flower Extract and their Antibacterial, Photocatalytic Activity. IJIRSET, 2016; 5(4): 6437- 6445.
- Baskar G, Chandhuru J, Fahad K S, Praveen SA. Mycological synthesis, characterization and antifungal Activity of Zinc Oxide Nanoparticles. Asian J Pharm Tech, 2013; 3(4): 142-146.
- Wani A H, Shah M A. A unique and profound effect of MgO and ZnO nanoparticles on some plant pathogenic fungi. J Appl Pharm Sci, 2012; 2(3): 40-44.
- Weidenkaffa A, Rellera W A, Wokaunb A, Steinfeldc A. Thermogravimetric analysis of the ZnO/Zn water splitting cycle. Thermochimica Acta, 2000; 359(1): 69-75.
- Poorna A C, Pradeep S N. Identification of novel chitinolytic Streptomyces. spp from a sacred grove and it’s in vitro antagonistic activity analysis. Int J Curr Microbiol App Sci, 2016; 5 (8): 916-928.
- Rana S, Salam DM. Antimicrobial potential of Actinomycetes isolated from soil samples of Punjab. India J Microbiol Exp, 2014; 1(2): 10.
- Mabrouk M I, Saleh N M. Molecular identification and characterization of antimicrobial active Actinomycetes strains from some Egyptian soils. American-Eurasian J Agric Environ Sci, 2014; 14(10): 954-963.
- Maleki H, Dehnad A, Hanifian S, Khani S. Isolation and molecular identification of Streptomyces spp. with antibacterial activity from northwest of Iran. Bioimpacts, 2013; 3(3): 129-134.
- Sambrook J, Fritsch E D, Maniatis T. 1989. Molecular cloning: a laboratory manual, pp. 49-55. 2nd Ed. Cold Spring Harbor Laboratory Press.
- Jadhav SR, Karad DD, Kulakrni WS. Isolation, identification and characterization of Keratinolytic Streptomyces coelicoflavus. Int J Curr Microbiol App Sci, 2016; 5(7): 153-163.
- Hashem M A, El-Tayeb N W, Ragab M Y, Fathy O D. Molecular identification of Streptomyces clavuligerus and structure elucidation of its antimicrobial efficiency. IJAPSA, 2016; 2: 96-105.
- Cregut M, Piutti S, Slezack-Deschaumes S, Benizri E. Compartmentalization and regulation of arylsulfatase activities in Streptomyces sp., Microbacterium sp. and Rhodococcus sp. soil isolates in response to inorganic sulfate limitation. Microbiol Res, 2013; 168(1): 12-21.
- Quiros M L, Hernandez C, Salas A J. Purification and characterization of an extracellular enzyme from Streptomyces antibioticus that converts inactive glycosylated oleandomycin into the active antibiotic. Eur J Biochem, 1994; 222(1): 129-135.
- Bhasin S, Modi A H. Optimization of fermentation medium for the production of glucose isomerase using Streptomyces sp. SB-P1. Biotechnol Res Int, 2012; 1-10 pages.
- Manhas K R, Bala S. Thermostable glucose isomerase production by some bacteria and Streptomyces. Indian J Microbiol, 2004; 44(2): 129-132.
- NevesAA, Pereira A D, Vieira M L, Menezes C J. Real time monitoring biomass concentration in Streptomyces claguligerus cultivations with industrial media using a capacitance probe. J Biotechnol, 2000; 84(1): 45-52.
- Mehrotra R S, Aggarwal A. 2013.Fundamentals of Plant Pathology, pp. 408 Mc Graw-Hill Education.
- Siddiqi S K, Husen A. Fabrication of metal nanoparticles from fungi and metal salts: scope and application. Nano Res Lett, 2016; 11:98.
- Kakhaki M Z, Youzbashi A, Naderi N. Optical Properties of Zinc Oxide nanoparticles prepared by a one-step mechanochemical synthesis method. J PhysSci, 2015; 26(2): 41-51.
- Gupta A, Srivastava P, Bahadur L, Amalnerkar D P, Chauhan R. Comparison of physical and electrochemical properties of ZnO prepared via different surfactant-assisted precipitation routes. Appl Nanosci, 2014; 5(7): 787-794.
- Saitou N,Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987; 4(6): 406-425.
- Jyotia M, Vijaya D, Radhac S. To study the role of temperature and sodium hydroxide concentration in the synthesis of Zinc Oxide nanoparticles. Int J Sci Res Pub, 2013; 3(11): 1-5.
- Hafez E E, Hassan S H, Elkady F M, Salama E. Assessment of antibacterial activity for synthesized zinc oxide nanorods against plant pathogenic strains. Int J Sci Tech Res, 2014; 3(9): 318-324.
- Ummartyotin S, Tangnorawich B. Data on the growth of ZnO nanorods on Nylon 6 and photocatalytic activity. Data in Brief, 2016; 8: 643-647.
- Navale RG, Thripuranthaka M, Late J D, Shinde SS. Antimicrobial activity of ZnO nanoparticles against pathogenic bacteria and fungi. JSM Nanotechnol Nanomed, 2013; 3(1): 1033.
- Chauhan R, Reddy A, Abraham J. Biosynthesis of silver and zinc oxide nanoparticles using Pichia fermentans JA2 and their antimicrobial property. Appl Nanosci, 2015; 5 (1): 63-71.
- Gunalana S, Sivaraja R,Rajendran V. Green synthesized ZnO nanoparticles against bacterial and fungal pathogens. Prog Nat Sci Mater Int, 2012; 22(6): 693-700.
- He L, Liu Y, Mustapha A, Lin M. Antifungal activity of zinc oxide nanoparticles against Botrytis cinerea and Penicillium expansum. Microbiol Res, 2011; 166(3): 207-215.
- Pinto M G, Nazareth R. Green synthesis and characterization of zinc oxide nanoparticles. J Chem Pharm Res, 2016; 8(6): 427-432.
- Wentzel A, Bruheim P, Verby A, Jakobsen M Y, Sletta H, Omara A M W, Hodgson A D, Ellingsen E T. Optimized submerged batch fermentation strategy for systems scale studies of metabolic switching in Streptomyces coelicolor A3(2). BMC Sys Biol, 2012; 6(1):59-74.
- EL-Moslamy S H. Large-Scale Biogenic Fabrication, Characterization, and Antibacterial Efficacy of High-Yield CuONPs Using Endophytic Streptomyces albus E56. Nanosci Nanotechnol Lett, 2018; 10(7): 933-942.
- Zahn J A. Scale-up and Optimization of Natural Product Fermentation Processes using Mass-Guided Metabolite Fingerprinting. Adv Biotech Micro, 2017; 3(3): 555614.
- Chakravarty I, Kundu K, Ojha1 S, Kund S. Development of Various Processing Strategies for New Generation Antibiotics Using Different Modes of Bioreactors. JSM Biotechnol Bioeng, 2017; 4(1): 1073.
- Yehia R S, Ahmed OF. In vitro study of the antifungal efficacy of zinc oxide nanoparticles against Fusarium oxysporum and Penicilium expansum. Afr J Microbiol Res, 2013; 7(19): 1917-1923.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.