ISSN: 0973-7510
E-ISSN: 2581-690X
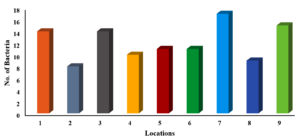
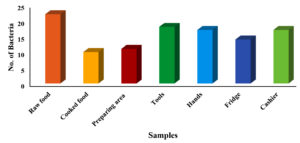
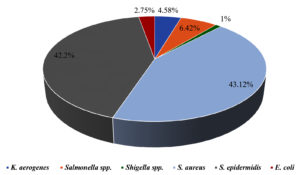
Foodborne illnesses are a major public health concern, and meat products are one of the most common sources of contamination. Handling and processing raw meat in restaurants can increase the risk of foodborne illnesses if the correct hygiene and safety measures are not followed. Consequently, it is important to conduct a comprehensive assessment of foodborne illness-causing microorganisms to monitor the food safety practices in restaurants and prevent the spread of contamination, protecting public health and ensuring the safety of the food supply chain. In view of this, this study conducted an assessment of local restaurants to identify the prevalence of different foodborne illness-causing microorganisms. A total of 63 samples were collected aseptically using cotton swabs from restaurants in 9 different locations in Al-Mandaq City, Saudi Arabia (7 samples from each location). Klebsiella aerogenes (K. aerogenes), Salmonella spp., Shigella spp., Staphylococcus aureus (S. aureus), Staphylococcus epidermidis (S. epidermidis) and Escherichia coli (E. coli) were isolated and identified from each sample using different media. From 63 samples, 91 isolates of pathogenic bacteria were isolated from 9 restaurants. Higher prevalence was found in location 7, where the number of isolates was 17, while the lowest pathogenic load was observed in location 2, where the number of isolates was 8. Among the samples, the highest number of pathogenic isolates was observed in raw foods (22), followed by samples collected from the tools (18). Of the 109 bacterial counts, S. aureus contributed 43.12%, followed by S. epidermidis (42.2%), Salmonella spp. (6.42%), and K. aerogenes (4.58%). The frequency of E. coli occurrence was low (2.75%) in all the samples collected from the nine locations.
Foodborne, Contamination, Pathogens, Bacteria, Detection, Staphylococcus
Foodborne illnesses are among the world’s top concerns in the twenty-first century.1,2 This issue is more significant in developing countries because of inadequate infrastructure and low levels of knowledge.3 Animal-derived foods intended for human consumption are typically the most dangerous if the correct food hygiene practices are not followed. In terms of pathogen matters, organic poisons, many potential pollutants, contaminants, and animal goods – including meats, fish, and their products – are typically classified as high-risk commodities.4 Enterohemorrhagic Escherichia coli (E. coli) O157 is one of the main pathogenic bacteria frequently linked to foods of animal origin. Hemolytic-uremic syndrome (HUS) and/or severe diarrhea are also possible outcomes of this pathogen’s asymptomatic disease in individuals.5-6 E. coli O157 infections in humans have primarily been linked to ingesting tainted, inadequately cooked minced beef and unpasteurized cow’s milk. Restaurants and butcher shops are regularly blamed for human infections caused by E. coli O157.7
Clostridium, Staphylococcus, Salmonella, Bacillus, Moraxella spp., Campylobacter, Listeria, Pseudomonas spp., lactic acid bacteria, and Acinetobacter spp. are the most prevalent genera of meat pathogenic bacteria.8 and can result in discoloration, foul odors, and slime on beef surfaces. Cladosporium, Geotrichum, Penicillium, and Mucor are some of the mold and yeast species that can be detected in meat, along with Candida and Cryptococcus species.9 During preparation and/or slaughter, the meat may have picked up pathogens. Most of the infecting organisms come from animal excrement and hide. Food of animal origin, including meats, fish, and their products, particularly when consumed uncooked, are typically viewed as high-risk foods.10-11
Meat serves as the primary source of protein and critical vitamins for most individuals in many parts of the world, and these are necessary for daily activities and the development, maintenance, and repair of body cells.12 Despite its nutritional content, fresh meat is quite susceptible to contamination. Consuming contaminated food might result in hospitalization, mild to severe disease, or death.5 Previous research from emerging and established nations has shown that foodborne illnesses might affect at least 10% of the population.13
Compared to other foodborne infections, the prevalence of Salmonella is still very high and has not decreased in over ten years.14 Salmonella is a primary contributor to several cases of foodborne illness and is occasionally one of the pathogens with the highest rates of morbidity and fatality.15
Numerous studies have shown that bacteria can adhere to stainless steel and other surfaces that come into contact with meat.16 Listeria monocytogenes has the highest hospitalization rates of all known foodborne pathogens and can potentially cause a severe and even fatal illness.17 Moreover, its attachment to processing equipment and the environment can cause critical issues.
Water activity and an ideal pH for fresh meat are considered to be key factors in the growth of microorganisms. Meat is therefore regarded as a food that spoils quickly.18 Cross-contamination between meat products and raw meats happens on butcher shop surfaces, as well as during processing, preparation, handling, and supplying afterward.19 Unsanitary handling techniques, the use of contaminated water and blades and other cutting equipment during cutting operations, and infectious panels to exhibit meat destined for sale lead to meat poisoning at slaughterhouses and retail meat shops. Knives, weighing scales, and wooden boards from retail stores can be contaminated with germs, especially those of the Shigella and Staphylococcus aureus (S. aureus) species.20
Meat bacterial counts are utilized as a reliable estimate of how hygienic meat is. Meat with a high bacterial load results from poor slaughterhouse infrastructure, filthy animals, and careless handling of carcasses. Therefore, evaluating the bacterial load can determine the threat to individual well-being.21 Many nations have advocated using sanitary and value management procedures for red meat and its products, particularly in food-serving. The environment in the butcher’s and abattoir areas can act as a health hazard if there is not sufficient hygiene control.20,22
The methods of managing and handling meat used in various restaurants might give rise to the possibility of a large number of spoilage germs easily growing on it and causing spoiling and foodborne illness. Therefore, it is important to evaluate the meat’s microbiological quality to develop sanitary procedures that can be utilized in butcheries to reduce the risk of contracting a foodborne illness from consuming tainted goods.10 Moreover, it is considered that water plays a crucial role in food safety in restaurants and industries. Water can be used to wash working surfaces, carcasses, equipment, employees’ hands, blood off meat, and much more. The characteristics of the water applied in meat processing in restaurants also have a significant role in preventing or increasing meat contamination.23
No similar data has been provided concerning the evaluation of meat safety practices, meat-related illnesses, and the microbiological load of meat cutter surfaces in Al-Mandaq City’s restaurants. This might make it more difficult for the government to implement policies that accurately address the adverse effects of meat infection issues on general health.24 In light of this, this study aimed to establish the needed database regarding the potential prevalence of different pathogenic bacteria in restaurants in Al Mandaq City, Saudi Arabia.
Sample collection
Abiotic and biotic samples were collected from raw food (shish tawook, chicken kebob), cooked food (shish tawook, chicken kebob, fries), preparation area, tools (knife, tweezers, board), hands, fridge, and cashier in restaurants in different locations in Al Mandaq City, which is in the southwestern region of Saudi Arabia. A total of 63 samples were collected aseptically using cotton swabs from 9 restaurants (7 samples from each location). Collected samples were stored in an icebox and transferred to the lab in sterilized containers.
Isolation and identification of bacterial strains
Swab streaking was done on various media following the protocol of Melebari et al.22 The media used were Eosin methylene blue (EMB) agar, HiCrome Staph Selective Agar, MacConkey agar, and blood agar, and they were incubated for 24 hours at 37°C. Morphologically distinct colonies were selected and purified after 24 hours of incubation. E. coli strains were identified using an EMB medium by producing a metallic sheen. Staphylococcus epidermidis (S. epidermis) and S. aureus were identified using HiCrome Staph Selective Agar. S. epidermidis and S. aureus were identified, based on the production of blue and green colonies on the media. Shigella spp. and Salmonella spp. were identified on Salmonella–Shigella agar.
Basic confirmation tests
As basic confirmation tests, catalase activity was tested, and gram staining of isolated strains was performed. The protocol of Smith and Hussey25 was used to perform gram staining. The catalase activity of the isolated strains was tested on the bacterium colony with 3% H2O2, and bubble formation was observed. The isolated strains’ hemolytic potential was tested on blood agar (nutrient agar + 10% sheep blood). Pathogens were streaked on blood agar and incubated for 24 hours at 37oC. The clear zone and color of colonies were used to check for hemolysis.
Statistical analysis
Data was represented using percentages and frequencies. SPSS 26 was used to analyze the data statistically. The Chi-square test was performed to compare the groups, and the significant impact was assessed at the 95% confidence interval (a = 5%) (Table 2 and 3).
The bacteria isolated from nine different restaurants were characterized and confirmed by gram staining, catalase, and hemolytic potential of the isolated strains. Table 1 shows that the Staphylococcus species are gram-negative, whereas all the other characterized strains are gram-positive. Only Klebsiella aerogenes (K. aerogenes) is catalase positive among the isolated bacterial strains, while the rest are negative. All the strains showed hemolytic activity except Salmonella spp.
Table (1):
Characterization of the isolated pathogens from different locations in Al-Mandaq City, Saudi Arabia
Strain |
Gram staining |
Catalase |
Hemolytic activity |
|---|---|---|---|
K. aerogenes |
– |
– |
+ |
S. aureus |
+ |
+ |
+ |
S. epidermidis |
+ |
+ |
+ |
Salmonella spp. |
– |
+ |
– |
Shigella spp. |
– |
+ |
+ |
E. coli |
– |
+ |
+ |
Prevalence of pathogens
Statistical analysis performed to determine the association between the presence of pathogens in different locations showed that there was a significant correlation between the location of the restaurant and K. aerogenes, Salmonella spp., S. aureus, and S. epidermidis, where it was observed that the p-value was greater than 0.05. A non-significant correlation was observed in Shigella spp. and E. coli, where it was observed that the p-value was greater than 0.05 ( Table 2). Pearson correlation of pathogens observed and sample type – raw food, cooked food, preparation area, tools (knife, tweezers, board), hands, fridge, and cashier – showed that there was no significant association (p > 0.05) except Salmonella spp., where a significant association was observed (p < 0.05).
Table (2):
Chi-square analysis of the isolated pathogens from all the locations
| K. aerogenes | Salmonella spp. | Shigella spp. | S. aureus | S. epidermidis | E. coli | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | |
| Pearson Chi-Square | 16.076a | 8 | .041 | 19.607a | 8 | .012 | 8.129a | 8 | .421 | 16.253a | 8 | .039 | 15.146a | 8 | .056 | 5.214a | 6 | .517 |
| Likelihood Ratio | 13.885 | 8 | .085 | 19.090 | 8 | .014 | 4.529 | 8 | .807 | 20.223 | 8 | .010 | 19.926 | 8 | .011 | 6.558 | 6 | .364 |
| Linear-by-Linear Association | .032 | 1 | .858 | 6.073 | 1 | .014 | .600 | 1 | .439 | 1.781 | 1 | .182 | .297 | 1 | .586 | .481 | 1 | .488 |
| N of Valid Cases | 63 | 63 | 63 | 63 | 63 | 63 | ||||||||||||
| a. 9 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .56. | a. 9 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .78. | a. 9 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .11. | a. 9 Cells (50.0%) have an expected count lower than 5. The minimum expected count is 1.78. | a. 9 Cells (50.0%) have an expected count lower than 5. The minimum expected count is 1.89. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .71. | |||||||||||||
Table (3):
Chi-square analysis of the isolated pathogens from all the samples
| K. aerogenes | Salmonella spp. | Shigella spp. | S. aureus | S. epidermidis | E. coli | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | Value | df | P-value | |
| Pearson Chi-Square | 5.214a | 6 | .517 | 11.250a | 6 | .081 | 6.097a | 6 | .412 | 7.875a | 6 | .247 | 4.350a | 6 | .629 | 9.100a | 6 | .168 |
| Likelihood Ratio | 6.558 | 6 | .364 | 13.426 | 6 | .037 | 3.991 | 6 | .678 | 9.862 | 6 | .131 | 4.322 | 6 | .633 | 8.308 | 6 | .216 |
| Linear-by-Linear Association | .481 | 1 | .488 | .356 | 1 | .551 | 2.250 | 1 | .134 | 2.494 | 1 | .114 | .178 | 1 | .673 | 3.100 | 1 | .078 |
| N of Valid Cases | 63 | 63 | 63 | 63 | 63 | 63 | ||||||||||||
| a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .71. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is 1.00. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .14. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is 2.29. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is 2.43. | a. 7 Cells (50.0%) have an expected count lower than 5. The minimum expected count is .43. | |||||||||||||
A total of 109 bacterial strains were isolated from 9 restaurants in Al-Mandaq City, Saudi Arabia. Among all the tools used, the highest number of pathogenic isolates was observed for location 7, where the total number of pathogens was 17, followed by location 9, where the number was 15. The same number of pathogens (14) was observed for location 1 and location 3. Similarly, for location 5 and location 6, an equal number of pathogens (11) was observed. A smaller number of pathogens (8) was recovered from location 2 (Figure 1).
The highest number of pathogens was recovered from raw foods (22), followed by tool samples (18). An equal load of pathogens was isolated from hands and cashier samples (17 each). The number of pathogens in the cooked food was relatively low (10) compared to the number of pathogens isolated from the other samples (Figure 2).
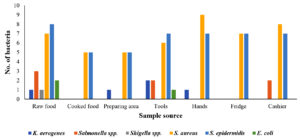
From 7 sources of each location, a total of 109 pathogens were isolated. From raw food, 22 pathogens were recovered, among which the most prevalent was S. epidermidis with 8 isolates, followed by S. aureus (n = 7) and Salmonella spp. (n = 3). Both K. aerogenes and Shigella spp. were isolated only once each. Furthermore, only S. aureus (n = 5) and S. epidermidis (n = 5) were recovered from cooked food. K. aerogenes was found in the preparation area (n = 1), while there was a greater abundance of S. aureus (n = 5) and S. epidermidis (n = 5) compared to the rest of the pathogens. From tools, K. aerogenes and Salmonella spp. were found in equal isolations (n = 2), whereas Shigella spp. was not observed. The highest number of isolations (n = 7) of S. epidermidis was observed from all the locations, followed by S. aureus (n = 6). No Salmonella spp. and Shigella spp. were observed from the hands. However, the highest number of S. aureus isolates was 9, and these were isolated from all the locations collectively. There were no K. aerogenes, Salmonella spp., E. coli, or Shigella spp. from the samples collected from the fridge. However, the number of S. aureus and S. epidermidis isolates (7 isolates) was found to be higher than that of the other pathogens. From cashier samples, S. aureus was found to be the pathogen with the highest number of isolates (8 isolates), followed by S. epidermidis (7 isolates), and there were no isolations of K. aerogenes, E. coli, or Shigella spp. (Figure 3). Figure 4 summarizes the percentage prevalence of the characterized pathogens. Of the 109 bacterial counts, S. aureus contributed 43.12%, followed by S. epidermidis (42.2%), Salmonella spp. (6.42%), and K. aerogenes (4.58%). The frequency of E. coli occurrence was very low (2.75%) in all the samples collected from the nine locations.
It has been shown that there is a daily increase in the prevalence of foodborne infections. Foodborne illnesses continue to be a major health concern for people worldwide and pose a threat to communities, the food industry, and individuals.26 Foodborne bacterial infections can be caused by the consumption of both raw and prepared foods/cooked foods; even ready-to-eat foods may be a source. A study conducted by Alharbi et al.27 revealed the isolation of pathogenic bacteria (Klebsiella pneumoniae, E. coli, S. aureus, and Bacillus cereus (B. cereus) from ready-to-eat foods.
According to Kirk et al., the most common bacterial pathogens that might cause foodborne diseases are Salmonella spp., E. coli, S. aureus, S. epidermidis, Campylobacter spp., and B. cereus.28 In general, foodborne infections are linked with gastroenteritis, which may be mild to acute in terms of severity.22 These infections produce symptoms such as diarrhea, nausea, and vomiting, which disturb various physiological functions by affecting the body’s main systems, including the cardiovascular, respiratory, musculoskeletal, and immune systems.29
The present study describes the prevalence of pathogenic bacteria isolated from restaurants in Al-Mandaq City, Saudi Arabia. It is clear from Figure 2 that most of the pathogens were isolated from raw food samples. This may be due to the contamination during transportation and processing,30 particularly the processing of meat, which can be easily contaminated by the tools used during cutting and preparation.31 The results of our study are consistent with the findings of Bantawa et al., who reported the isolation of S. aureus, Salmonella spp., and Shigella spp. from raw foods.32 In cooked food, the rate of the existence of S. aureus and S. epidermidis was alarmingly high, as 83.32% of samples were positive, and these were the most common pathogens found in all samples. It is well known that S. aureus and S. epidermidis can be destroyed by heat.33 In the current study, the possible reason for their presence in cooked food is the contaminated surface that comes in contact with cooked foods, naked hands, or other contaminated tools.34
Salmonella spp. was isolated in 6.42% of samples from raw food, cashier, and the tools used for cutting and processing (Figure 4). With a slight variation, our results align with those of Arumugaswamy et al.,35 who reported the presence of Salmonella spp. in samples from raw foods and cooked foods, whereas in our study, we did not observe the presence of this pathogen in cooked food. Shigella spp. was recovered only from raw food in 1% of all the samples collected from all the locations. Shigellosis is a worldwide public health issue caused by members of the genus Shigella. Previous studies have focused on human gastrointestinal pathogens but ignored animal groups. Shigella spp. infects and causes clinical symptoms in animals, and these animals act as a source of Shigella spp.36
The second highest pathogenic population was observed in the tools (Figure 2), such as knives, cutting boards, gloves, storage, and other instruments used to process foods, which are potential sources of contamination. Gurmu and Gebretinsae reported that this high load of pathogenic bacteria might be due to the persistence of the pathogens on the surface of these tools.17 It should also be noted that either the workers or their clothing may be the source of contamination.20 In our study, we only isolated K. aerogenes from raw foods, food preparation areas, tools, and hands of the workers. Klebsiella species are found in soil, water, and food sources, as well as on the mucosal surfaces of mammals.37
With a slight variation, our results align with the findings of Hemalata and Virupakshaiah,38 who reported the isolation of pathogenic populations from different food sources, among which the highest prevalence was observed for Pseudomonas (23%), followed by S. aureus (22%), Salmonella spp. (21%), E. coli (22.32%), and Klebsiella spp. (9%). The contradiction with this study is that we found a very low E. coli population (2.75%) from all the samples. It is concluded that either the environment was unsuitable for E. coli, there was no cross-contamination, or the water served in the restaurant was E. coli-free since one of the main sources of E. coli contamination is water.39 In another study, it was found that Salmonella species, C. perfringens, S. aureus, and other foodborne pathogenic bacterial species are frequently present in foods.40
Six types of pathogens with various frequencies were isolated from nine different restaurants in Al-Mandaq City, Saudi Arabia. Of all the restaurants, the bacterial load was highest in location 7, and raw food samples were more contaminated than the others. The most abundant pathogens were S. aureus (43.12%) and S. epidermidis (42.2%), and the least abundant pathogen was Shigella spp.
Utilization of such contaminated foods, particularly cooked food, is a dilemma for the people of the city that should be addressed with proper checks and balances by the government. Future studies must include other advanced techniques such as antimicrobial susceptibility testing and molecular identification of all the isolated strain-based 16S rRNA genes to determine the exact characterizations and prevalence of various pathogens.
ACKNOWLEDGMENTS
Author would like to thank Dr. Tahir Taha and Mr. Tariq Alpakistany for their assistance with collecting the samples, and for their comments that greatly improved the manuscript. Also, would like to thank Al-Baha University for providing the greatest lab environment and facilities.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Trevisan C, Torgerson PR, Robertson LJ. Foodborne Parasites in Europe: Present Status and Future Trends. Trends Parasitol. 2019;35(9):695-703.
Crossref - Khan JA, Rathore RS, Ahmad I, et al. Assessment of Foodborne Bacterial Pathogens in Buffalo Raw Milk Using Polymerase Chain Reaction Based Assay. Food-borne Pathogens and Disease. 2022;19(11):750-757.
Crossref - Scapini V. Disaster, Infrastructure Damage, and Health. Reconstruction. 2020;1(2):219-225.
Crossref - Bintsis T. Foodborne pathogens. AIMS microbiology, 2017; 3(3), 529–563.
Crossref - Bannon J, Melebari M, Jordao Jr C, Leon-Velarde CG, Warriner K. Incidence of top 6 Shiga toxigenic Escherichia coli within two Ontario beef processing facilities: challenges in screening and confirmation testing. Aims Microbiol. 2016;2(3):278-291.
Crossref - Samreen, Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers: a potential threat to public health. Journal of Global Antimicrobial Resistance. 2021;27:101-111.
Crossref - Beyi AF, Fite AT, Tora E, et al. Prevalence and antimicrobial susceptibility of Escherichia coli O157 in beef at butcher shops and restaurants in central Ethiopia. BMC Microbiol. 2017;17:1-6.
Crossref - Anas M, Ahmad S, Malik A. Microbial Escalation in Meat and Meat Products and Its Consequences. In A. Malik, Z. Erginkaya, & H. Erten (Eds.), Health and Safety Aspects of Food Processing Technologies. Springer International Publishing. 2019:29-49.
Crossref - Sevik R, Akarca G, Kilinc M, Ascioglu C. Chemical Composition of Tea Tree (Melaleuca alternifolia) (Maiden & Betche) Cheel Essential Oil and Its Antifungal Effect on Foodborne Molds Isolated from Meat Products. Journal of Essential Oil Bearing Plants. 2021;24(3):561-570.
Crossref - Atlabachew T, Mamo J. Microbiological quality of meat and swabs from contact surface in butcher shops in Debre Berhan, Ethiopia. J Food Qual. 2021;7520882.
Crossref - Arafa SH, Alshehri WA, Organji SR, et al. Antimicrobial Resistance, Virulence Factor-Encoding Genes, and Biofilm-Forming Ability of Community-Associated Uropathogenic in Western Saudi Arabia. Pol J Microbiol. 2022;71(3):325-339.
Crossref - Akram M, Munir N, Daniyal M, Egbuna C, Gaman M-A, Onyekere PF, Olatunde A. Vitamins and Minerals: Types, Sources and their Functions. In C. Egbuna & G. Dable Tupas (Eds.), Functional Foods and Nutraceuticals: Bioactive Components, Formulations and Innovations. Springer International Publishing. 2020:149-172.
Crossref - Zerabruk K, Retta N, Muleta D, Tefera AT. Assessment of microbiological safety and quality of minced meat and meat contact surfaces in selected butcher shops of Addis Ababa, Ethiopia. J Food Qual. 2019;3902690.
Crossref - Hurd HS, Enoe C, Sorensen L, et al. Risk-Based Analysis of the Danish Pork Salmonella Program: Past and Future. Risk Analysis. 2008;28(2):341-351.
Crossref - Ho-Palma AC, Gonzales-Gustavson E, Quispe E, et al. Salmonella in Chicken and Pork Meat are Likely to be a Major Contributor to Foodborne Illness in Peru. Heliyon. 2022.
Crossref - Evans A, Slate AJ, Akhidime ID, Verran J, Kelly PJ, Whitehead KA. The Removal of Meat Exudate and Escherichia coli from Stainless Steel and Titanium Surfaces with Irregular and Regular Linear Topographies. Int J Environ Res Public Health. 2021;18(6):3198.
Crossref - Hadjilouka A, Loizou K, Apostolou T, Dougiakis L, Inglezakis A, Tsaltas D. Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor. Biosensors. 2020;10(11):178.
Crossref - Verma AK, Chatli MK, Mehta N, Kumar P. Antimicrobial and Antioxidant Potential of Papain Liver Hydrolysate in Meat Emulsion Model at Chilling Storage Under Aerobic Packaging Condition. Waste and Biomass Valorization. 2022;13(1):417-429.
Crossref - Barril PA, Soto SA, Jaureguiberry MV, et al. Microbiological risk characterization in butcher shops from the province of Neuquen, Patagonia Argentina. LWT. 2019;107:35-40.
Crossref - Bersisa A, Tulu D, Negera C. Investigation of bacteriological quality of meat from abattoir and butcher shops in Bishoftu, Central Ethiopia. Int J Microbiol.;2019:6416803.
Crossref - Azwai S, Alfallani E, Abolghait S, et al. Isolation and Molecular Identification of Vibrio spp. by Sequencing of 16S rDNA from Seafood, Meat and Meat Products in Libya: A Descriptive Study. Innovations in Microbiology and Biotechnology. 2022;4:104-114.
Crossref - Melebari M, Alpakistany T, Taha TM, Alsalman AS. Prevalence of pathogenic bacteria on face masks from wet markets in Makkah duringthe COVID-19 pandemic. The Scientific Journal of King Faisal University: Basic and Applied Sciences, 2022; 23(2), 33–8.
Crossref - Rani ZT, Mhlongo LC, Hugo A. Microbial Profiles of Meat at Different Stages of the Distribution Chain from the Abattoir to Retail Outlets. Int J Environ Res Public Health. 2023;20(3).
Crossref - Gurmu E, Gebretinsae H. Assessment of Bacteriological Quality of Meat Cutting surfaces in selected Butcher shops of Mekelle city, Ethiopia. Journal of Environmental and Occupational Science. 2013;2:1.
Crossref - Smith AC, Hussey MA. Gram stain protocols. American Society for Microbiology. 2005;1:14.
- Devleesschauwer B, Haagsma JA, Mangen M-JJ, Lake RJ, Havelaar AH. The Global Burden of Foodborne Disease. In T. Roberts (Ed.), Food Safety Economics: Incentives for a Safer Food Supply. Springer International Publishing. 2018:107-122.
Crossref - Alharbi SA, Abdel-Ghaffar MH, Kadher NR. Isolation and identification of pathogenic bacteria from ready-to-eat fast foods in Al-Quwayiyah, Kingdom of Saudi Arabia. Afr J Food Agric Nutr Dev. 2019;19(3):1439-1451.
Crossref - Kirk MD, Pires SM, Black RE, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12(12):e1001921.
Crossref - Akhtar S, Sarker MR, Hossain A. Microbiological food safety: a dilemma of developing societies. Crit Rev Microbiol. 2014;40(4):348-359.
Crossref - Ercolini D, Russo F, Torrieri E, Masi P, Villani F. Changes in the Spoilage-Related Microbiota of Beef during Refrigerated Storage under Different Packaging Conditions. Appl Environ Microbiol. 2006;72(7):4663-4671.
Crossref - Eisel WG, Linton RH, Muriana PM. A survey of microbial levels for incoming raw beef, environmental sources, and ground beef in a red meat processing plant. Food Microbiol. 1997;14(3):273-282.
Crossref - Bantawa K, Rai K, Subba Limbu D, Khanal H. Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC Res Notes. 2018;11(1):618.
Crossref - Strunk T, Richmond P, Prosser A, Simmer K, Levy O, Burgner D, Currie A. Method of bacterial killing differentially affects the human innate immune response to Staphylococcus epidermidis. Innate Immunity. 2011;17(6):508-516.
Crossref - Syne SM, Ramsubhag A, Adesiyun AA. Microbiological hazard analysis of ready-to-eat meats processed at a food plant in Trinidad, West Indies. Infect Ecol Epidemiol. 2013;3(1):20450.
Crossref - Arumugaswamy RK, Rusul G, Abdul Hamid SN, Cheah CT. Prevalence of Salmonella in raw and cooked foods in Malaysia. Food Microbiol. 1995;12:3-8.
Crossref - Zhu Z, Wang W, Cao M, et al. Virulence factors and molecular characteristics of Shigella flexneri isolated from calves with diarrhea. BMC Microbiol. 2021;21(1):214.
Crossref - Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9(4):228-236.
Crossref - Hemalata V, Virupakshaiah D. Isolation and identification of food borne pathogens from spoiled food samples. Int J Curr Microbiol Appl Sci. 2016;5(6):1017-1025.
Crossref - Oje O, Ajibade VA, Fajilade OT, Ajenifuja O. Microbiological analysis of ready-To-eat (RTE) foods vended in mobile outlet catering units from Nigeria. J Adv Food Sci Technol. 2018;5(1):15-19.
- Muleta D, Ashenafi M. Salmonella, Shigella and growth potential of other food-borne pathogens in Ethiopian street vended foods. East African Medical Journal. 2001;78(11):576-580.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.