ISSN: 0973-7510
E-ISSN: 2581-690X
It has been established that the urinary tract is not sterile; however, research related to the study of urinary bacteria is limited. This study aimed to investigate the frequency and patterns of resistance of normal urinary aerobic bacterial flora and clean catch midstream urine specimens collected from 120 young healthy females and cultured. Bacterial identification and antimicrobial susceptibility were performed using the Biomérieux VITEK® 2 automated system. Participants who had undergone antimicrobial treatment within one month were not included. The incidence of positive bacterial cultures was 54.2%, of which 21.5% were polymicrobial. Approximately 107 bacterial isolates that encompass 12 genera and 27 species that were predominated by gram-positive bacteria (72%) were cultivated. Staphylococcaceae (46.1%) and Enterobacteriaceae (17.8%) were the most frequent isolates among gram-positive and gram-negative bacteria, respectively, of which 36 species have been identified as β-lactamase producers. The top four frequently isolated bacteria were Micrococcus spp. (16%), Staphylococcus haemolyticus (13.2%), Staphylococcus aureus (10%), and Klebsiella pneumoniae (10%). Twenty-two bacterial species were subjected to antimicrobial susceptibility testing using broad- and narrow-spectrum antibiotics and antimicrobials, which showed the lowest susceptibility rate against gram-positive bacteria, followed by erythromycin and azithromycin. A lower antimicrobial susceptibility potential among gram-negative bacteria was observed against ampicillin, followed by piperacillin and cefotaxime. Our findings emphasize the importance of highlighting urine bacterial flora in studies, especially those related to susceptibility patterns, by employing more advanced culture methods as multiple drug-resistant bacteria were isolated.
Urine Flora, Urinary Tract Infection Susceptibility Test, Biomerieux VITEK® 2 System, Multidrug Resistance
Recent studies have emphasized the presence of bacteria in the urine of women; the fact decided that the use of antibiotics based on a positive culture should be accompanied by clinical symptoms.1,2 For the time being, culture methods were regarded by some microbiologists as ineffective in reflecting bacterial diversity in the human body, as are some technological methods that were developed after the tremendous progress in genomics, which has led to an unprecedented revolution in deep recognition of body flora.3 However, despite their limitations in identifying many types of bacteria, as well as the fact that they are accused of being the cause of ancient prevailing concept, which states that the urinary tract is not occupied naturally by bacteria, notwithstanding, the culture methods remain with all strength at a number that cannot be exceeded due to their importance in identifying patterns of bacterial susceptibility to antimicrobial agents, which represents the initial phase in setting up suggestions for powerful treatment of infectious diseases.4,5 Therefore, unsurprisingly, Robert Koch considered them fundamental to infectious disease studies.6 Under usual conditions, normal flora do not pose a danger to the host, but recent studies have reported that these bacteria possessed or can extrinsically acquire some genes responsible for resistance to some antibiotics, which has sounded the alarm bell for this emergency and drew attention to the necessity of conducting susceptibility testing for them.7-9 After tracing and extrapolating the study that focused on bacterial flora, it became clear to us that information on susceptibility patterns of these microorganisms is very limited, especially those that settle in the urinary tract. Therefore, this study was conducted to investigate the urinary flora in young, healthy females and to assess the antibiotic susceptibility of recovered bacteria.
Study population
Participants enrolled in the study were unmarried Saudi females between 21-23 years of age recruited at the Princess Nourah bint Abdulrahman University and King Khalid Hospital (n=120). The females had no history of overactive bladder syndrome, urinary incontinence, urgency, or frequency.
Study area and specimen collection
The study was conducted at the Princess Nourah Bint Abdulrahman University and King Saud University Research Centre in Riyadh. After collection of the urine sample, the midstream clean catch urine specimen was aliquoted into sterile 15 mL conical tubes and stored at −20°C.
Ethical approval
All procedures and techniques used in this study were in accordance with national regulations that govern the protection of human participants. The study protocols were approved by the Princess Nourah Bint Abdulrahman Institutional Review Board ( IRB number:17-0060). Urine specimens were collected after obtaining signed informed consent from all the participants. All the participants were >16 years of age.
Specimen culture and bacterial identification
Urine culture was performed by inoculating 200 µL of urine onto the entire blood agar plate. The plates were then incubated overnight at 37°C under aerobic conditions. Identification of bacteria and antibiotic susceptibility were carried out using the Biomerieux VITEK® 2.
Data analysis
Data were analyzed using SPSS (version 21) and Microsoft Excel 2010 software. Descriptive statistics were performed for the numbers of susceptible and resistant bacterial species, and the results are expressed as mean ± standard deviation. The Kruskal-Wallis test was used to determine statistical differences between the means of antibiotic resistance. The Mann-Whitney U test was performed for each pair of antibiotics and bacteria to determine the cause of the difference. The mean difference was significant at the level of 0.05.
There were 120 female participants recruited at the Princess Nourah bint Abdulrahman University and King Khalid Hospital. In the 20–25-year-old age group, none of them showed signs or symptoms of urinary tract infection. It was observed that 45.8% of the investigated midstream urine specimens were sterile. Positive cultures were obtained from 65 (54.2%) specimens, of which 14 were ploymicrobial. One hundred and seven bacterial isolates that encompass 12 genera and 27 species that were predominated by gram-positive bacteria, which represent 72%, were cultivated. Staphylococcus (46.1%) and Enterobacteriaceae (17.8%) were the most frequent isolates among the gram-positive and gram-negative bacteria, respectively. The frequency, percentage, and number of isolated urinary floras are presented in Tables 1 and 2. Of the 10 isolates, two Staphylococcus aureus strains were identified as methicillin-resistant Staphylococcus aureus (MRSA); however, they remained susceptible to vancomycin. It is worth noting that 36 β-lactamase-producing gram-positive bacteria were identified. Twenty-two bacterial species were subjected to antimicrobial susceptibility testing using broad- and narrow-spectrum antibiotics and antimicrobials. The mean numbers of susceptible, relatively resistant, and resistant species among gram-negative bacteria were 1.19 ± 0.81, 1.19 ± 1.25, and 3.86 ± 2.17, respectively. In contrast, the mean number of susceptible, relatively resistant, and resistant species among gram-positive bacteria were 2.63 ± 1.92, 3.68 ± 2.26, and 6.42 ± 3.49, respectively.
Table (1):
The profile, percentage and numbers of the isolated Gram-positive bacteria.
Genus |
Species |
No. of the isolates |
(%) |
|---|---|---|---|
Enterococcus |
Enterococcus faecalis |
3 |
2.8 |
Enterococcus faecium |
3 |
2.8 |
|
Kocuria |
Kocuria kristinae |
1 |
0.9 |
Micrococcus |
Micrococcus |
17 |
16 |
Rothia |
Rothia dentocariosa |
3 |
2.8 |
Rothia mucilaginosa |
1 |
0.9 |
|
Staphylococcus |
Staphylococcus aureus |
10 |
9.4 |
Staphylococcus auricularis |
2 |
1.9 |
|
Staphylococcus cohnii |
1 |
0.9 |
|
Staphylococcus epidermidis |
6 |
5.7 |
|
Staphylococcus haemolyticus |
14 |
13.2 |
|
Staphylococcus hominis |
2 |
1.9 |
|
Staphylococcus hyicus |
1 |
0.9 |
|
Staphylococcus intermedius |
2 |
1.9 |
|
Staphylococcus lugdunensis |
3 |
2.8 |
|
Staphylococcus sciuri |
6 |
5.7 |
|
Staphylococcus simulans |
1 |
0.9 |
|
Staphylococcus warneri |
1 |
0.9 |
Table (2):
The profile, percentage, and numbers of the isolated Gram-negative bacteria.
Genus |
Species |
No. of the isolates |
(%) |
|---|---|---|---|
Acinetobacter |
Acinetobacter baumannii |
2 |
1.9 |
Cadecea |
Cedecea lapagei |
1 |
0.9 |
Citrobacter |
Citrobacter murliniae |
1 |
0.9 |
Escherichia |
Escherichia coli |
7 |
6.6 |
Klebsiella |
Klebsiella pneumoniae |
10 |
9.4 |
Pseudomonas |
Pseudomonas aeruginosa |
5 |
4.7 |
Pseudomonas fluorescens |
1 |
0.9 |
|
Pseudomonas stutzeri |
1 |
0.9 |
|
Burkholderia |
Burkholderia cepacia |
1 |
0.9 |
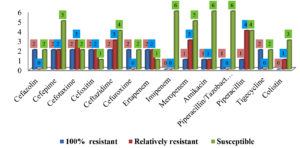
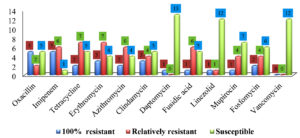
Figures 1 and 2 describe the numbers and patterns of susceptibility of gram-positive and gram-negative bacteria to antibiotics used for each species. Table 3 shows the patterns of susceptibility to seven antibiotics simultaneously tested against both types of bacteria.
Table (3):
Susceptibility patterns to the antibiotics tested against both bacterial types.
| Antimicrobial agent | 100% resistant | Relatively resistant | Susceptible | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gram negative | Gram positive | Gram negative | Gram positive | Gram negative | Gram positive | Gram negative | Gram positive | |
| Amoxicillin/Clavulanic acid | 2 | 5 | 0 | 6 | 2 | 1 | 4 | 12 |
| Ampicillin | 0 | 8 | 1 | 3 | 3 | 3 | 4 | 14 |
| Gentamicin | 1 | 2 | 1 | 4 | 6 | 6 | 8 | 12 |
| Levofloxacin | 0 | 2 | 0 | 2 | 8 | 10 | 8 | 14 |
| Ciprofloxacin | 0 | 3 | 0 | 3 | 8 | 8 | 8 | 14 |
| Moxifloxacin | 1 | 2 | 1 | 1 | 2 | 9 | 4 | 12 |
| Sulfamethoxazole/Trimethoprim | 1 | 2 | 1 | 4 | 3 | 6 | 5 | 12 |
Ampicillin showed the lowest susceptibility against gram-positive bacteria, followed by erythromycin and azithromycin. High susceptibility rates were reported for vancomycin (0%), daptomycin (1.3%), and linezolid (2.6%). The susceptibility profiles of gram-positive bacteria to the 19 antibiotic agents are summarized in Table 4.
Table (4):
Susceptibility profile of Gram-positive bacterial flora.
| Species | Antibiotics | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | OXA | IPM | CIP | LVX | MXF | TET | SXT | ERY | AZM | CLI | DAP | GEN | FUS | LZD | MUP | FOS | VAN | |
| E. faecalis(3) | – | 0 | – | – | 0 | 0 | – | 66.7 | – | 66.7 | – | – | 0 | – | – | 0 | – | – | – |
| E. faecium(3) | – | 0 | – | – | 100 | 0 | – | 0 | – | 100 | – | – | 0 | – | – | 33.3 | – | – | – |
| K. kristinae(1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Micrococcus spp.(17) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| R. dentocariosa(3) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| R. mucilaginosa(1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| S. aureus(10) | 50 | 80 | 50 | 50 | 10 | 0 | 0 | 50 | 20 | 60 | 70 | 50 | 0 | 10 | 50 | 0 | 30 | 20 | 0 |
| S. auricularis(2) | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 50 | 0 | 50 | 50 | 50 | 0 | 0 | 0 | 0 | 50 | 50 | 0 |
| S. cohnii(1) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 |
| S. epidermidis(6) | 33.3 | 100 | 33.3 | 33.3 | 0 | 0 | 0 | 33.3 | 0 | 83.3 | 66.7 | 16.7 | 0 | 0 | 16.7 | 0 | 0 | 0 | 0 |
| S. haemolyticus(14) | 64.3 | 71.4 | 64.3 | 64.3 | 7.1 | 7.1 | 7.1 | 57.1 | 14.3 | 71.4 | 71.4 | 21.4 | 0 | 7.1 | 42.9 | 0 | 7.1 | 14.3 | 0 |
| S. hominis(2) | 50 | 100 | 100 | 50 | 0 | 0 | 0 | 0 | 0 | 50 | 50 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 0 |
| S. hyicus(1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 |
| S. intermedius(2) | 50 | 50 | 50 | 50 | 0 | 0 | 0 | 50 | 50 | 50 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. lugdunensis(3) | 33.3 | 100 | 33.3 | 33.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33.3 | 0 | 0 | 0 | 0 | 0 |
| S. sciuri(6) | 100 | 100 | 100 | 100 | 16.7 | 50 | 0 | 50 | 16.7 | 100 | 100 | 100 | 0 | 83.3 | 33.3 | 0 | 16.7 | 16.7 | 0 |
| S. simulans(1) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 |
| S. warneri(1) | 100 | 100 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 39 | 53.2 | 39 | 39 | 10.4 | 7.8 | 3.9 | 31.2 | 10.4 | 46.8 | 40.3 | 23.4 | 1.3 | 13 | 20.8 | 2.6 | 9.1 | 10.4 | 0 |
AMC Amoxicillin/clavulanic acid, AMP Ampicillin, OXA Oxacillin, IPM Imipenem, CIP Ciprofloxacin, LVX Levofloxacin, MXF Moxifloxacin, TET Tetracycline, SXT Sulfamethoxazole/trimethoprim, ERY Erythromycin, AZM Azithromycin, CLI Clindamycin, DAP Daptomycin, GEN Gentamycin, FUS Fusidic acid, LZD Linezolid, MUP Mupirocin, FOS Fosfomycin, VAN Vancomycin
The susceptibility profile of gram-negative bacteria to 22 antibiotic agents is summarized in Table 5. A lower antimicrobial potential was observed against ampicillin, followed by piperacillin and cefotaxime. Ciprofloxacin and levofloxacin remained active with a resistance rate of 0%.
Table (5):
Susceptibility profile of Gram-negative bacterial flora.
| Species | Antibiotics | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | CFZ | CFP | CTX | FOX | CAZ | CXM | ETP | IPM | MEM | AMK | GEN | LFX | CIP | MXF | TZP | PIP | TET | TGC | SXT | CST | ||
| A. baumannii(2) | – | – | – | 0 | 50 | – | 0 | – | – | – | 0 | 0 | 0 | 0 | 0 | – | – | 50 | 0 | – | 0 | – | |
| C. lapagei(1) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | – | |
| C. murliniae(1) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | – | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 0 | – | |
| E. coli(7) | 0 | 14.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14.3 | 0 | 14.3 | 14.3 | 0 | 0 | – | |
| K. pneumoniae(10) | 0 | 100 | 0 | 0 | 0 | 10 | 10 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 20 | 0 | 10 | 0 | |
| P. aeruginosa(5) | – | – | – | 20 | 60 | – | 20 | – | – | 0 | 20 | 20 | 20 | 0 | 0 | – | 0 | 0 | – | – | – | 20 | |
| P. fluorescens(1) | – | – | – | 0 | – | – | 0 | – | – | 0 | 100 | 0 | 0 | 0 | 0 | – | 0 | 0 | – | – | – | 0 | |
| P. stutzeri(1) | – | – | – | 0 | – | – | 0 | – | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | – | – | – | 0 | |
| B. cepacia(1) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| Total | 6.9% | 44.8% | 6.9% | 10.3% | 20.7% | 10.3% | 13.8% | 6.9% | 10.3% | 0.0% | 10.3% | 6.9% | 6.9% | 0.0% | 0.0% | 6.9% | 3.4% | 24.1% | 17.2% | 6.9% | 6.9% | 3.4% | |
AMC Amoxicillin/Clavulanic acid, AMP Ampicillin, CFZ Cefazolin, CFP Cefepime, CTX Cefotaxime, FOX Cefoxitin, CAZ Ceftazidime, CXM Cefuroxime, ETP Ertapenem, IPM Imipenem, MEM Meropenem, AMK Amikacin, GEN Gentamicin, LFX Levofloxacin, CIP Ciprofloxacin, MXF Moxifloxacin, TZP Piperacillin/Tazobactam, PIP Piperacillin, TET Tetracycline, TGC Tigecycline, SXT Sulfamethoxazole/Trimethoprim, CST Colistin
Natural bacteria in the urinary tract have not been extensively studied, similar to skin and gut bacteria and not included in the human microbiome project,4 study on their antimicrobial susceptibility patterns have not obtained enough, which may be because the urinary system was formerly deemed to be a sterile niche.10,11 However, recent studies have shown that the urinary tract is naturally colonized by a variety of bacterial species,12 and it has become certain that their lop-sidedness somehow have a role in the system’s physiology and susceptibility to infection, as they represent a biological defence line against “pathogen induced inflammation”.13,14 Numerous studies have confirmed differences in bacterial communities between healthy women and women with urgency urinary incontinence (UUI).15,16 Thomas et al and Mueller et al correlated response to treatment in patients with UUI with dysbiosis of the urinary flora.17,18
In the present study, antibiotics did not undergo susceptibility tests against Micrococci, Rothiae, Kocuriakristinae, and Burkholderiaceae. This is because automated identification systems, although they allow accurate identification, unfortunately do not perform susceptibility tests for some bacterial species, which remains one of the most important deficiencies of these systems.19,20 There are no adequate recent studies to indicate the pattern of micrococcal response to antibiotics, but it is convenient here to refer to the Dürstetal findings that showed the response of Micrococcus luteus to penicillin.21 Scarce information is available on the pathogenesis of Micrococcus spp. as well, but in general, Micrococcus spp. seldom produce β-lactamases,22 and it is difficult to attribute the etiology of the infection to this genus if it is isolated from a clinical specimen, as it rarely affects people with a healthy immune system. The high virulence remains limited to people with low immunity, such as Acquired immunodeficiency syndrome (AIDS) patients.23,24 Nevertheless, the high virulence have given rise to infections, such as pneumonia, meningitis, septic arthritis, peritonitis, and endophthalmitis.25-29 We isolated 5 genera out of the six multiple drug resistant (MDR) bacteria, which are abbreviated by “ESKAPE” using their acronyms as follows: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.30
Approximately 14.3% of Escherichia coli isolates were resistant to ampicillin, piperacillin, tetracycline, and moxifloxacin; however, they were inhibited by penicillin agents combined with β-lactamase inhibitors (amoxicillin/cavulanic acid [amoxiclav] and piperacillin/tazobactam [TZP]). None of the isolates were resistant to imipenem, tigecycline, or colistin, similar to the findings of Rasheed et al.,31 who reported resistance to ampicillin and tetracycline. However, they documented resistance to cefotaxime (5.3%), gentamycin, and amoxicillin/cavulanic acid (4.6 %), which is in agreement with our results, where no resistance was encountered, as the above-mentioned antibiotics. Of 11 antibiotics, Acinetobacter baumannii isolates were susceptible to 9 antibiotics and only 50% of the isolates were resistant to cefotaxime and piperacillin, and these outcomes are good as the bacterium has emerged as an opportunistic MDR pathogen associated with nosocomial infections32; however, to best of our knowledge, no literature was available with regards to the susceptibility of urinary Acinetobacter baumannii flora. All Klebsiella pneumoniae strains were resistant to ampicillin and susceptible to amoxiclav. This might suggest that the mechanism of resistance may involve TEM β-lactamases, an enzyme that mediates ampicillin resistance, and can be inhibited by β-lactamase inhibitors,33 which is similar reason that can explain the susceptibility of Klebsiella pneumoniae isolates to TZP. Osman et al. contradicted our results when they reported colistin and ampicillin resistance among Klebsiella pneumoniae strains isolated from milk; however, similar to the recent outcomes, there was no resistance to gentamycin, cefatoxime, and sulfamethoxazole/trimethoprim.34 One strain exhibited resistance to ertapenem and meropenem, which is alarming because the genes encoding Klebsiella pneumoniae carbapenemase and New Delhi metallo β-lactamase are harbored in plasmids; thus, can be transmitted to other species.35 The susceptibility patterns of Pseudomonas fluorescens and Pseudomonas stutzeri were found to be similar, except that the former was resistant to meropenem. Kittinger et al. reinforced our susceptibility results for meropenem against Pseudomonas fluorescens; however, they reported resistance to ceftazidime and ciprofloxacin.36 Pseudomonas stutzeri was susceptible to 19 antibiotics tested identically to its susceptibility results when isolated from a patient with peritonitis.37 Park et al. attributed the high susceptibility of this bacterium to its rare clinical incidence with less exposure to antimicrobials.37 Of 21 isolates, Cedecea lapagei was resistant to 14 antibiotics, including amikacin, gentamycin, ceftazidime, cefepime, and sulfamethoxazole/trimethoprim, an outcome contradicted with that of Biswal et al., who reported bacterial susceptibility to the above-mentioned antibiotics.38 This may provide a prime example of bacterial adaptation to antibiotics through the acquisition of resistance determinants.33,39 However, our findings are in agreement with those of Biswal et al., who demonstrated bacterial susceptibility to meropenem and ciprofloxacin along with resistance to tetracycline and tigecycline.38 Citrobacter murliniae was identified as an operational taxonomic unit in female urine in 2017,40 but to our knowledge, no literature is available about its susceptibility profile. Citrobacter murliniae expressed a MDR phenotype similar to that of Cedecea lapagei (Table 5). The two enterococci were susceptible to ampicillin, levofloxacin, and daptomycin. A similar percentage of penicillin (96%) was reported by Rudy et al. in their screening of Enterococcus faecalis isolated from urine, but they observed a higher resistance rate to ciprofloxacin (43%).41 They found that 14% of the Enterococcus faecium strains were resistant to ciprofloxacin, 32% to ampicillin, and 19% to tetracycline. Our study is consistent with previous studies, showing that the urinary tract is predominantly inhabited by coagulase-negative staphylococci (CONS).42,43 Staphylococcus haemolyticus was documented as a part of the normal female urinary flora42; however, Pindar et al. reported the organism’s susceptibility to vancomycin.44
Staphylococcus simulans was found to be resistant to all β-lactams and quinolones. Staphylococcus simulans was isolated by Shields et al. as a skin-associated pathogen45; however, a few data are available about the antimicrobial susceptibility of this emerging pathogen, and we did not recover Staphylococcus saprophyticus, although it is associated with community-acquired urinary tract infections, secondary to Escherichia coli.43,46 To best of our knowledge, it was not isolated from midstream urine except in low percentages, ranging between 2-4% in sexually active females and pregnant women,47-50 where it was encountered in low bacteriuria.51 The difference in the prevalence percentages can also be attributed to the fact that the prevalence of Staphylococcus saprophyticus changes seasonally and increases significantly at the end of summer.52 We isolated three coagulase-positive Staphylococci: Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus hyicus (which were found to be MDR), but it is surprising that the second and third are related to dogs and pigs, respectively,53,54 and there is no evidence. To the best of our knowledge, there is no evidence that this bacterium is part of the natural components of the human urinary system. Staphylococcus aureus exhibited a significantly lower susceptibility (P≤0.05) to tetracycline, erythromycin, azithromycin, clindamycin, mupirocin, and fusidic acid than CONS. However, the latter demonstrated a higher resistance rate to the remaining antibiotics, which is consistent with the results reported by Tao et al.55 Compared to CONS with a resistance rate average of 20.3%, Staphylococcus aureus showed lower resistance rate of 0.0–10% to quinolone. Likewise, resistance to vancomycin and linezolid was not observed among Staphylococcus aureus isolates. This also emphasizes compatibility with the findings of Tao et al.
Our study showed that the number of isolated aerobic bacterial flora in clean catch midstream urine was significantly higher in gram-positive bacteria. This is a good indication that gram-positive bacteria, including MRSA and other β-lactamase producers do not show any resistance to vancomycin. Imipenem, levofloxacin, and ciprofloxacin were completely effective against gram-negative bacteria. However, it is concerning that resistance to some antibiotics is mediated by mobile genetic elements that can be transmitted between bacterial species. Therefore, these results are not conclusive, rather we recommend more studies in this field by employing modern methods of culture, such as matrix-assisted laser desorption ionization-time of flight mass spectrometry, to isolate broader spectra of bacteria, ensuring a deeper study of susceptibility and resistance patterns of urine bacterial flora.
ACKNOWLEDGMENTS
The authors would like to thank Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project No. (PNURSP2022R31).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study is approved by the Institutional Review Board, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia (IRB number:17-0060).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: Use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(3):871-876.
Crossref - Price TK, Dune T, Hilt EE, et al. The clinical urine culture: Enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54(5):1216-1222.
Crossref - Lagier J-C, Hugon P, Khelaifia S, Fournier P-E, Scola B La, Raoult D. The Rebirth of Culture in Microbiology through the Example of Culturomics To Study Human. Gut Microbiota. 2015;28(1):237-64.
Crossref - Wolfe AJ, Brubaker L. Sterile Urine and the Presence of Bacteria. Eur Urol. 2015:173-174.
Crossref - Graf EH, Simmon KE, Tardif KD, et al. Unbiased Detection of Respiratory Viruses by Use of RNA Sequencing-Based Metagenomics: a Systematic Comparison to a Commercial PCR Panel. J Clin Microbiol. 1000;54(4):1000-1007.
Crossref - Lagier J-C, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin Microbiol Rev. 2015;28(1):208-236.
Crossref - Sabino YNV, Santana MF, Oyama LB, et al. Characterization of antibiotic resistance genes in the species of the rumen microbiota. Nat Commun. 2019;10(1):5252.
Crossref - Langelier C, Graves M, Kalantar K, et al. Microbiome and antimicrobial resistance gene dynamics in international travelers. Emerg Infect Dis. 2019;25(7):1380-1383.
Crossref - Burcham ZM, Schmidt CJ, Pechal JL, et al. Detection of critical antibiotic resistance genes through routine microbiome surveillance. PLoS One. 2019;14(3):e0213280.
Crossref - Karstens L, Asquith M, Caruso V, et al. Community profiling of the urinary microbiota: considerations for low-biomass samples. Nat Rev Urol. 2018;15(12):735-749.
Crossref - Aragon IM, Herrera-Imbroda B, Queipo-Ortuno MI, et al. The Urinary Tract Microbiome in Health and Disease. Eur Urol Focus. 2018;4(1):128-138.
Crossref - Magistro G, Stief CG. The Urinary Tract Microbiome: The Answer to All Our Open Questions? Eur Urol Focus. 20195(1):36-38.
Crossref - Kline KA, Lewis AL. Gram-Positive Uropathogens, Polymicrobial Urinary Tract Infection, and the Emerging Microbiota of the Urinary Tract. Microbiol Spectr. 2016;4(2):10.
Crossref - Nienhouse V, Gao X, Dong Q, et al. Interplay between Bladder Microbiota and Urinary Antimicrobial Peptides: Mechanisms for Human Urinary Tract Infection Risk and Symptom Severity. PLoS One. 2014;9(12):e114185.
Crossref - Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. MBio. 2014;5(4):1283-1297.
Crossref - Karstens L, Asquith M, Davin S, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol. 2016;6:78.
Crossref - Thomas-White KJ, Hilt EE, Fok C, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J. 2016;27(5):723-733.
Crossref - Mueller ER, Wolfe AJ, Brubaker L. Female urinary microbiota. Curr Opin Urol. 2017;27(3):282-286.
Crossref - Kandi V, Palange P, Vaish R, et al. Emerging Bacterial Infection: Identification and Clinical Significance of Kocuria Species. Cureus. 2016.
Crossref - Dotis J, Printza N, Stabouli S, Papachristou F. Kocuria species peritonitis: Although rare, we have to care. Perit Dial Int. 2015;35(1):26-30.
Crossref - Kanjanauthai S, Kanluen T. Propionibacterium acnes: A rare cause of late prosthetic valve endocarditis and aortic root abscess. Int J Cardiol. 2008;130(2):e66-e68.
Crossref - Szczerba I. Susceptibility to antibiotics of bacteria from genera Micrococcus, Kocuria, Nesterenkonia, Kytococcus and Dermacoccus. Med Dosw Mikrobiol. 2003;55(1):75-80. PMID: 12908418
- The Prokaryotes: A Handbook on the Biology of Bacteria by Martin M. Dworkin. https://www.goodreads.com/book/show/11219882-the-prokaryotes. Accessed March 15, 2021.
- Smith, Neafie, Yeager, Skelton. Micrococcus folliculitis in HIV-1 disease. Br J Dermatol. 1999;141(3):558-61.
Crossref - Salar A, Carratala J, Fernandez-Sevilla A, Marin D, Granena A. Pneumonia caused by Micrococcus species in a neutropenic patient with acute leukemia [2]. Eur J Clin Microbiol Infect Dis. 1997:546-548.
Crossref - Fosse T, Toga B, Peloux Y, Granthil C, Bertrando J, Sethian M. Meningitis due to Micrococcus luteus. Infection. 1985;13(6):280-281.
Crossref - Wharton M, Rice JR, McCallum R, Gallis HA. eptic arthritis due to Micrococcus luteus. J Rheumatol. 1986 Jun;13(3):659-660. PMID: 3735296
- Kao CC, Chiang CK, Huang JW. Micrococcus species-related peritonitis in patients receiving peritoneal dialysis. Int Urol Nephrol. 2014;46(1):261-264.
Crossref - Cartwright MJ, King MH, Weinberg RS, Guerry RK. Micrococcus Endophthalmitis. Arch Ophthalmol. 1990:1523-1524.
Crossref - Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J Infect Dis. 2008;197(8):1079-1081.
Crossref - Rasheed MU, Thajuddin N, Ahamed P, Teklemariam Z, Jamil K. Resistencia microbiana a drogas em linhagens de Escherichia coli isoladas de fontes alimentares. Rev Inst Med Trop Sao Paulo. 2014;56(4):341-346.
Crossref - Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii An emerging opportunistic pathogen. Virulence. 2012;3(3):243-250.
Crossref - Hoffman SB. Mechanisms of Antibiotic Resistance. Compend Contin Educ Pract Vet. 2001;23(5):464-472.
Crossref - Osman KM, Hassan HM, Orabi A, Abdelhafez AST. Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathog Glob Health. 2014;108(4):191-199.
Crossref - Yan J, Pu S, Jia X, et al. Multidrug resistance mechanisms of Carbapenem resistant Klebsiella pneumoniae strains isolated in Chongqing, China. Ann Lab Med. 2017;37(5):398-407.
Crossref - Kittinger C, Lipp M, Baumert R, et al. Antibiotic resistance patterns of Pseudomonas spp. isolated from the river Danube. Front Microbiol. 2016;7:586.
Crossref - Park SW, Back JH, Lee SW, et al. Successful antibiotic treatment of Pseudomonas stutzeri-induced peritonitis without peritoneal dialysis catheter removal in continuous ambulatory peritoneal dialysis. Kidney Res Clin Pract. 2013;32(2):81-83.
Crossref - Biswal I, Hussain N, Grover R. Cedecea lapagei in a patient with malignancy: Report of a rare case. J Cancer Res Ther. 2015;11(3):646.
Crossref - Liebl W, Kloos WE, Ludwig W. Plasmid-borne macrolide resistance in Micrococcus luteus. Microbiology. 2002;148(8):2479-2487.
Crossref - Gottschick C, Deng ZL, Vital M, et al. The urinary microbiota of men and women and its changes in women during bacterial vaginosis and antibiotic treatment. Microbiome. 2017;5(1):99.
Crossref - Rudy M, Nowakowska M, Wiechula B, Zientara M, Radosz-Komoniewska H. Antibiotic susceptibility analysis of Enterococcus spp. isolated from urine. Przegl Lek. 2004;61(5):473-476. PMID: 15515808
- Marrie TJ, Kwan C, Noble MA, West A, Duffield L. Staphylococcus saprophyticus as a cause of urinary tract infections. J Clin Microbiol. 1982;16(3):427-431.
Crossref - Anderson JD, Clarke AM, Anderson ME, Isaac-Renton JL, McLoughlin MG. Urinary tract infections due to Staphylococcus saprophyticus biotype 3. Can Med Assoc J. 1981;124(4):415-418.
- Pindar C, Viau RA. Staphylococcus haemolyticus epididymo-orchitis and bacteraemia: a case report. JMM Case Reports. 2018;5(7):e005157.
Crossref - Shields BE, Tschetter AJ, Wanat KA. Staphylococcus simulans: An emerging cutaneous pathogen. JAAD Case Reports. 2016;2(6):428-429.
Crossref - Hovelius B, Mardh PA. Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev Infect Dis. 1984;6(3):328-337.
Crossref - Ehlers S, Merrill SA. Staphylococcus Saprophyticus. 2022 Jun 27. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan. PMID: 29493989.
- Goldenring JM. Urinary tract infection with Staphylococcus saprophyticus. J Adolesc Heal Care. 1986;7(6):417-418.
Crossref - Hedman P, Ringertz O. Urinary tract infections caused by Staphylococcus saprophyticus. A matched case control study. J Infect. 1991;23(2):145-153.
Crossref - Sousa VS De, Da-Silva APDS, Sorenson L, et al. Staphylococcus saprophyticus recovered from humans, food, and recreational waters in Rio de Janeiro, Brazil. Int J Microbiol. 2017;2017:4287547.
Crossref - Rupp ME, Soper DE, Archer’ GL. Colonization of the Female Genital Tract with Staphylococcus saprophyticus. J Clin Microbiol. 1992;30(11).
Crossref - Galinski J, Namysl E. Role of Staphylococcus saprophyticus in urinary infections. Pol Tyg Lek. 1991;46(40-42):746-749. PMID: 1669150.
- (PDF) Coagulase positive Staphylococcal colonization of humans and their household pets. https://www.researchgate.net/publication/40042655_Coagulase_positive_Staphylococca
_colonization_of_humans_and_their_household_pets. Accessed March 18, 2021. - Casanova C, Iselin L, Von Steiger N, Droz S, Sendi P. Staphylococcus hyicus bacteremia in a farmer. J Clin Microbiol. 2011;49(12):4377-4378.
Crossref - Tao H, Wang J, Li L, Zhang HZ, Chen MP, Li L. Incidence and antimicrobial sensitivity profiles of normal conjunctiva bacterial flora in the central area of China: A hospital-based study. Front Physiol. 2017;8:363.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.