ISSN: 0973-7510
E-ISSN: 2581-690X
The objective of this study was to isolate and select Lactobacillus strains with probiotic features for a potential use as starter for the preservation of animal feed. Olives, cow’s and camel’s milk, butter, beer drech and maize silage were used as isolation sources. Molecular identification using 16S rRNA gene sequencing identified four isolates of Lactobacillus plantarum and three of Lactobacillus fermentum. The different screening tests revealed that four strains: Lactobacillus plantarum OV13, Lactobacillus sp. OV15, Lactobacillus fermentum E161 and Lactobacillus sp. E631 are the best probiotic candidates, based on the results of their tolerance to acidic pH between 2 and 3.5, to 0.3% bile, to artificial gastric conditions for 18h of incubation, to their antibiotics resistance, their inhibition power on pathogenic bacteria (Pseudomonas sp., Staphylococcus aureus, Escherichia coli and Listeria ivanovii), and finally to their adhesion capacity to intestinal epithelial cells. The selected probiotic strains are considerate as good candidates for further investigation and should be tested, in vivo, to elucidate their potential health benefits on the animal performance as novel probiotic starters.
Probiotics, lactic acid bacteria, Lactobacillus, screening.
The fermented products are an important source of lactic acid bacteria (LAB). Some of these bacteria endowed with specific properties, act positively on human and animal health, and they are called probiotics.1 Probiotics are live microorganisms that can be supplemented to diverse types of products: food, drugs and food supplement in order to establish a beneficial gut microflora. Metchnikoff2 showed that probiotics offer health a bigger longevity. The species belong to Lactobacillus and Bifidobacterium are the most commonly used microorganisms in probiotic products. Some species of yeast like Saccharomyces cerevisiae and Saccharomyces boulardii and others species of E. coli and Bacillus are also used.3
Contrary to the chemicals complements, which can have toxic consequences on the body, probiotics allow the reconstitution of the natural conditions by remedying the deficiencies,4 fight the pathogenic bacteria by acting as alternative to antibiotics5,6 and so, prevent the gastrointestinal disorders and intestinal infections.6 Probiotics also have possibility to influence positively the immune system of the host6 by preventing a colorectal cancer using cell apoptosis mechanism.7
The selection of probiotic bacteria is a difficult task, because they have to resist the stressful conditions of gastrointestinal tract and adhere to the intestinal epithelium, to assure their survival in it, so that they can make their benefaction on the host8,9
In order to select LAB with probiotic potential, this study focused on the bacilli form of LAB isolates. Fourteen isolates of Lactobacillus sp. isolated from different fermented products were confronted to different tests to evaluate their probiotic proprieties.
Isolation of Lactic Acid Bacteria
Various samples were taken from different fermented products in west of Algeria (olives, cow’s and camel’s milk, butter, maize silage and beer drech), and used as bacterial sources. The dilution plate method, on acidified deMan Rogosa and Sharpe (MRS, pH 5.4) agar medium, was used to select LAB and promoting the selection of the bacilli form. Plates were incubated at 37°C for 24h to 48h under anaerobic conditions (anaerobic jar) and all candidates that corresponded to LAB appearance were selected and purified by sub culturing, then used to verify their probiotic capacity.
Molecular Identification of the Isolates
Total DNA Extraction
Total DNA was extracted from 2ml overnight cultures. After centrifugation at 12000g for 5min, cells were collected and washed twice, with sterile distilled water. The pellets were resuspended in 400µl of lysis buffer (2% Triton X-100, 10mM NaCl, 10mM Tris-HCl, pH 8, 1 mM EDTA, 3% SDS) containing lysozyme 3 mg/ml, and then incubated for 1 h at 37°C. Suspensions were grinded for 2min with 0.6g of sterile beads and 400µl of phenol /chloroforme (V/V). Phases were separated by centrifugation for 5 min at 12000g, then, in a new Eppendorf microtube, the aqueous phase was carefully mixed with 1 µl of RNase (10 mg/ml) and incubated for 15min at 37°C. Equal quantity of phenol/ chloroform was added to the aqueous phase, then, the mixture was vortexed and centrifuged for 5min at 12000g. The aqueous phase was transfered in another Eppendorf in order to precipitate DNA with 40µl of sodium acetate 3M pH 7 and 800µl of absolute cold ethanol with incubation for 30min on ice, followed by a centrifugation at 4°C for 10min at 12000g. DNA was washed with ethanol 70% and centrifuged at 12000g for 5min, then, dried during 5 min and resuspended in 50µl of Tris-EDTA. Finally, Total DNA was stored at –20°C.
PCR Amplification of 16S rRNA Gene
PCR amplification of the 16S rRNA gene was carried out using primers: BSF8-20 (5’-AGA GTT TGA TCC TGG CTC AG -3’) and BSR1541-20 (5’-AAG GAG GTG ATC CAG CCG CA- 3’), with a total volume of 50 ìl containing: 2.5µl of DNA, 25µl of Mix DNA enzyme (2X), 5µl of each primer, and 12.5µl of sterile water. PCR conditions were: initial DNA denaturation at 94°C for 2 min followed by 29 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s and elongation at 72°C for 1 min, then a final cycle at 72°C for 5 min. Amplicons were purified using Wizard SV Gel and PCR Clean-Up System and Sequencing. The purified PCR products were sequenced then the obtained sequences were compared to those in the GenBank database using the BLAST algorithm.
Acidity and Artificial Gastric Conditions Tolerance
Overnight cultures were centrifuged at 12000 g for 10min then the pellets were washed three times in sterile phosphate saline buffer (PBS) pH 7.4, and cultured on MRS agar pH 3.5. The viability of cells was recorded after 24h of incubation in anaerobic atmosphere at 37°C.
Isolates were tested for their capacity to grow after both 2h and 18h of incubation, on a broth simulating gastric conditions (NaCl 125 Mm/L, KCl 7 Mm/L, NaHCO3 45 Mm/L, pepsin 3 g/L). Overnight cultures were centrifuged at 12000 g for 15min at 4°C, then washed twice with phosphate buffer (50 Mm/L, pH 6.5), and resuspended in 3ml of the same buffer. One milliliter of this suspension was pelleted at 12000g for 5min at 4°C, and finally resuspended in 10 ml of artificial gastric solution. The final pH was adjusted to pH 2 and pH 3, and anaerobic conditions were maintained.10,11
Bile Tolerance
Two different methods were used to verify the strains resistance to bile
The first method consisted in inoculating bacteria on MRS broth supplemented with 0.3% natural bovine bile pH 4 for 24h at 37°C in anaerobic conditions. The same bacteria were inoculated for the second time on MRS agar pH 5.8 for 72h, to evaluate the tolerance on bile salt.12
In the second method, the isolates were inoculated in MRS broth (pH 4 supplemented with 0.3% of bile salt) and incubated at 37°C in anaerobic conditions, once for 3h, and once for 24h. The suspensions were re-suspended in MRS broth without bile (pH 5.8), to evaluate the survival percentage of LAB by the measure of the optical density (OD) at 600 nm.
The percentage of the growth was calculated as follows:
Survivalrate = OD of MRS with bile / OD of MRS without bile × 100
Bile resistance corresponds to a percentage superior to 50%.10,13
Antibiotic Susceptibility
To test isolates susceptibility to antibiotics, the disc diffusion method was employed. Overnight cultures were inoculated on Muller Hinton agar. After solidification, different antibiotic discs including: vancomycin (30µg), pristinamycin (15µg), nalidixic acid (30µg), oxacilin1 (1µg), rifampicin (30µg), streptomycin (10µg), penicillin (6µg), imipenem (10µg), cefazolin (30µg), piperacilin (75µg), gentamicin (10µg), cefsulodin (30µg) and colistin (50µg), were placed on the medium surface. The plates were then incubated at 37°C for 24h to 48h on anaerobic conditions. Results were expressed as sensitive (S) or resistant (R) according to the National Committee for Clinical Laboratory Standards14.
Antibacterial Activity
To detect the antibacterial activity, the LAB cultures were confronted to various pathogenic bacterial strains: Escherichia coli, Staphylococcus aureus (ATCC 43300), Staphylococcus aureus (ATCC 25923), Pseudomonas sp. and Listeria ivanovii. The LAB were inoculated using spot-on-the-lawn technique, on plates containing 15 ml of MRS agar, and then incubated at 37°C for 18h in anaerobic conditions. The pathogenic cultures used as indicators and inoculated in appropriate semi-liquid media at a rate of 1 % (Muller-Hinton for E. coli and Staphyloccocus, Brains Heart Infusion for Listeria and King A for Pseudomonas), were used to overlay the LAB colonies, and then incubated for the second time aerobically at 37°C. The antibacterial activity was determined by the presence of a clear zone around the colonies.15,16
In Vitro Adherence Test to Epithelial Cells
Intestinal Epithelial Cell Suspension
Chicken intestines were used for this assay. After washing them several times in the PBS buffer until clarification, the intestinal segment was opened and held in cold PBS 0.1 M, pH 7.2, supplemented with 1% penicillin-streptomycin for 1 h under shaking. Intestinal epithelial cells were collected by scraping the intestinal wall in Dulbecco’s Modified Eagle Medium (DMEM) pH 7 supplemented with 1% penicillin-streptomycin, and held for 30 min in this medium at 4°C under agitation. Cell suspensions were centrifuged at 6000 g for 10min, the pellet was washed three times in cold PBS pH 7, then resuspended in DMEM and conserved at 4°C.16,17
Bacterial Suspensions
Overnight cultures of LAB, were centrifuged at 12000g for 10 min and washed twice in sterilized cold PBS 0.1 M, pH 7.2, then held in the same buffer.
Adherence Assay
For adhesion assay, 1ml of epithelial cells (105 106 cell/ ml) was mixed with 1ml of bacteria cells (107 108 CFU/ ml) and incubated at 37°C for 24h. This mixture was pelleted at 6000 g for 10 min and washed three times with PBS then filtered on 0.45µm sterile filter to remove nonadherent bacteria. Finally, the filter was washed in 10ml of DMEM. The In vitro adhesion of LAB to epithelial cells was observed directly with photonic microscopy using oil immersion, after staining with methylen blue. The adherence was also examined by Analytic Scanning Electron Microscopy (JEOL JSM-6610LA) after dehydratation only in a graded series of ethanol, without fixation step16, 17.
Isolation and Identification of Isolates
Origins and identification of some isolates are shown in Table 1. Identified strains were divided into two groups. Using the Basic Local Alignment Search Tool in the NCBI site, four strains isolated from human food products, were identified as Lactobacillus plantarum with 100% homology and three strains isolated from silage belonged to Lactobacillus fermentum with 99% homology.
Table (1):
Origins of selected LAB and their identification.
Strain code |
Origin |
Identified as |
|---|---|---|
OV01 |
Fermented green olive |
Lactobacillus sp. |
OV13 |
Fermented green olive |
Lactobacillus plantarum |
OV15 |
Fermented green olive |
Lactobacillus sp. |
LV21 |
Fermented Cow milk |
Lactobacillus sp. |
OV60 |
Fermented purple olive |
Lactobacillus plantarum |
LC87 |
Fermented camel milk |
Lactobacillus sp. |
D006 |
Beer drech |
Lactobacillus plantarum |
B001 |
Butter |
Lactobacillus plantarum |
E161 |
Maize silage |
Lactobacillus fermentum |
E522 |
Maize silage |
Lactobacillus sp. |
E551 |
Maize silage |
Lactobacillus sp. |
E631 |
Maize silage |
Lactobacillus sp. |
E623 |
Maize silage |
Lactobacillus fermentum |
E652 |
Maize silage |
Lactobacillus fermentum |
Acidity and Simulated Gastric Conditions Tolerance
Results in table 2, show that all isolates have capacity to grow on acidic MRS agar (pH 3.5). In the simulated gastric solution, the survival was promising for all strains at pH 3 for 18h of incubation, but not for all, at pH 2. So, the majority of strains showed decreasing in their growth at pH 2 after 18h of incubation, whereas, strains E522 and D006 grew only after 2h and not after 18h of incubation; and three strains (E161, E551 and E623) did not grow in these conditions yet.
Table (2):
Acidity and simulated gastric conditions tolerance of selected LAB.
| MRS | Simulated gastric conditions | ||||
|---|---|---|---|---|---|
| pH 3.5 | pH 2 | pH 3 | |||
| Strain code | 2 h | 18 h | 2 h | 18 h | |
| OV01 | + | + | + | + | + |
| OV13 | + | + | + | + | + |
| OV15 | + | + | ± | + | + |
| LV21 | + | + | ± | + | + |
| OV60 | + | + | + | + | + |
| LC87 | + | + | ± | + | + |
| D006 | + | + | – | + | + |
| B001 | + | + | + | + | + |
| E161 | + | – | – | + | + |
| E522 | + | + | – | + | + |
| E551 | + | – | – | + | + |
| E623 | + | – | – | + | + |
| E631 | + | + | ± | + | + |
| E652 | + | + | ± | + | + |
+: presence of growth, – : absence of growth, ±: weak
Bile Tolerance
For bile tolerance, the majority of selected LAB grew in the presence of natural bovine bile, but they showed different behavior with bile salts, depending on the time of incubation. Thus, some strains (OV13, D006, and E522) which tolerated 0.3% bile salts after 3 h of incubation, their growth rate has been decreased after 24 h, contrary to the others (E161, E623 and E631) where the growth has been increased (table 3). Also, results showed that strains E551 and E652 did not grow in the presence of bovine bile.
Table (3):
Bile Tolerance of the selected LAB.
| Strain code | MRS with 0.3% bovine bile pH 4 | Percentage of tolerance to 0.3% salts bile pH 4 | |
|---|---|---|---|
| 3 h | 24 h | ||
| OV01 | + | 5.57±5.12 | 46.53±4.00 |
| OV13 | + | 75.12±1.45 | 53.28±16.88 |
| OV15 | + | 5.44±0.56 | 46.21±11.56 |
| LV21 | + | 10.55±0.88 | 13.92±4.77 |
| OV60 | + | 11.39±6.77 | 45.32±9.38 |
| LC87 | + | 42.76±13.41 | 30.20±9.32 |
| D006 | + | 83.15±8.85 | 52.89±7.74 |
| B001 | + | 7.77±0.33 | 47.58±9.91 |
| E161 | + | 24.51±5.54 | 59.98±8.35 |
| E522 | + | 70.12±4.28 | 47.08±10.90 |
| E551 | – | 1.97±1.19 | 25.85±5.38 |
| E623 | + | 5.95±0.9 | 59.12±9.50 |
| E631 | + | 5.79±1.95 | 56.40±4.97 |
| E652 | – | 2.78±0.15 | 28.43±5.41 |
+: presence of growth, – : absence of growth, ±: weak growth
each value in the table represents the mean value ± standard deviation from three trials
Antibiotic Susceptibility
The data obtained after screening for antibiotic resistance are shown in table 4. With the exception of some strains, the majority of them presented similar antibiotic profile. All the selected Lactobacilli strains were resistant to the nalidixic acid (30 µg) and vancomycin (30 µg), and are sensitive to imipenem (10 µg).
Table (4):
Antibiotic susceptibility of the selected LAB.
OV01 |
OV13 |
OV15 |
LV21 |
OV60 |
LC87 |
D006 |
B001 |
E161 |
E522 |
E551 |
E623 |
E631 |
E652 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
PT |
S |
S |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
S |
S |
NA |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
OX1 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
RA |
S |
S |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
S |
R |
S |
R |
R |
R |
R |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
P |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
R |
R |
R |
VA |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
IPM |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
CZ |
S |
S |
S |
S |
R |
S |
S |
R |
S |
S |
S |
R |
S |
R |
PIP |
S |
S |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
S |
S |
GM |
S |
R |
R |
R |
S |
R |
R |
R |
S |
R |
R |
S |
S |
R |
CFS |
S |
R |
S |
R |
S |
S |
S |
S |
S |
S |
S |
R |
R |
R |
CS |
S |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
PT: pristinamycin (15 µg), NA :nalidixicacid (30 µg), VA: vancomycin (30 µg) , OX1 :oxacilin1(1 µg),
RA: rifampicin (30 µg), S: streptomycin (10 µg), P: penicillin (6 µg), IPM: imipenem (10 µg), CZ: cefazolin (30 µg),
PIP: piperacilin (75 µg), GM: gentamicin (10 µg), CFS: cefsulodin (30 µg), CS: colistin (50 µg).
S: sensitive, R : resistant.
Antibacterial Activity
In this study, the antibacterial activity was detected with the majority of the selected strains, but with variations in their potential of inhibition. Results showed that the majority of strains have a good antimicrobial activity against Pseudomonas sp., Staphylococcus aureus (ATCC43300) and E. coli; and presented the lowest inhibition against Listeria ivanovii (table5). However, the nature of the inhibitive agent secreted by the strains must be identified later in further study.
Table (5):
Antibacterial activity of the selected LAB.
Strain code |
E. coli |
Staphylococcus Aureus ATCC25923 |
Staphylococcus Aureus ATCC43300 |
Listeria ivanovii |
Pseudomonas sp. |
|---|---|---|---|---|---|
OV01 |
11 |
7 |
24 |
– |
26 |
OV13 |
6 |
4 |
10 |
– |
16 |
OV15 |
18 |
11 |
15 |
12.5 |
+ |
LV21 |
16 |
9 |
12 |
± |
32 |
OV60 |
26 |
8 |
15 |
± |
24 |
LC87 |
8 |
5 |
12 |
± |
10 |
D006 |
– |
– |
– |
– |
– |
B001 |
14 |
8 |
16 |
– |
38 |
E161 |
5 |
5 |
9 |
– |
+ |
E522 |
5 |
7 |
11 |
– |
+ |
E551 |
– |
– |
– |
– |
– |
E623 |
11 |
– |
10 |
– |
14 |
E631 |
15 |
8 |
14 |
5 |
35 |
E652 |
4 |
– |
– |
– |
+ |
Diameters of inhibitions zones are expressed in mm.
– : no inhibition, diameter of inhibition zone d” 5mm: weak inhibition, diameter of inhibition zone Ã5mm: middle inhibition, + and diameter of inhibition zone e”15mm: strong inhibition.
Adhesion Capacity of Strains
The capacity of the selected strains to adhere to chicken’s epithelial cells was defined by light microscopy, using methylene-blue stain (Fig. 1a) and by the environmental scanning electron microscopy (Fig. 1b). Thus, six strains (OV13, OV15, OV60, D006, E161, E631) showed, In vitro, a good ability to adhere to intestinal epithelial chicken’s cells.
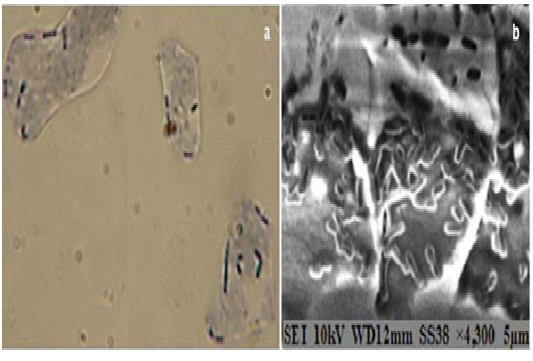 Fig. 1. Adhesion of isolated Lactobacillus on chicken epithelial cells: (a) Adhesion of Lactobacillus plantarum OV13 to the chicken epithelial cells as observed by methylen blue staining under light microscope (magnification 1000×); (b) Adhesion of Lactobacillus fermentum E161 as observed under electron microscopy (magnification 4300×)
Fig. 1. Adhesion of isolated Lactobacillus on chicken epithelial cells: (a) Adhesion of Lactobacillus plantarum OV13 to the chicken epithelial cells as observed by methylen blue staining under light microscope (magnification 1000×); (b) Adhesion of Lactobacillus fermentum E161 as observed under electron microscopy (magnification 4300×)In the present study, different Algerian fermented products were used as source of LAB. Some selected bacteria were identified using molecular methods and screened, in vitro, for probiotic proprieties such tolerance to gastrointestinal conditions, which are considered as one of the main factors limiting the use of microorganisms as live probiotic agents, as they determine their ability to survive in small intestine, and consequently, their capacity to play their functional role as probiotics18,19
The 16S rRNA gene sequencing of some isolates revealed two LAB species: L. plantarum (4 isolates) and L. fermentum (3 isolates). Results of the molecular identification are in agreement with data obtained by Khedid, et al.20, Jamaly, et al.10 and Mahmoudi, et al.,21 who showed the presence of L. plantarum species in dairy products, and with Kacem and Karam22 and Hurtado, et al.,23 who reported that L. plantarum and L. pentosus are the predominant species in fermented table olives; but are in disagreement with Yang, et al.,24 and Pang, et al.25 who did not reveal the presence of L. fermentum in silage. However, the potential of these bacterial species as a probiotic was previously reported in several studies.10,13,19
Survival of bacterial strains in simulated gastric conditions can be accurate indication of the ability of strains to survive passage through the host stomach. So, our results revealed that all selected bacteria were able to grow on acidic environment, between pH 2 and pH 3.5, depending on the strains, and all of them could survive after an exposure of 18 h on simulated gastric broth which contain 3g/L pepsin (pH 3). In fact, Gu, et al.26 and Argyri, et al. 27 mentionned similar data after 5 h of incubation in simulated gastrointestinal tract and Dunne, et al.28 reported that Lactobacilli strains can be resistant to pH varying between 2.5 and 4. However, the viability of some isolated LAB was affected at pH 2 and strains showed different behavior according to their time of incubation in this pH. Similar results were noted by Gu, et al.26, Pan, et al.9 and by Lahteinen, et al.29 in an acidic medium (pH 2) containing bile.
Ruiz, et al.18 mentioned that bacteria inhabiting intestinal tract must have intrinsic resistance mechanisms to cope with bile salts. So, our results showed that the majority of selected strains were able to tolerate 0.3% of bile, but had different behavior with the bovine and the chemical bile. In fact , Many studies have shown that LAB were able to resist to bile,30,31 and, it was observed , previously, by Dunne, et al.28 that some strains of Lactobacillus and Bifidobacterium had a better capacity of growth on human bile than on chemical bovine and porcine bile.
According to these first results, we suggested that some bacterial strains need time of adaptation to survive in hostile environment, and considered that the strains which could survive, in vitro, in the acidic gastric conditions and the bile, can survive, in vivo, in the digestive tract26, and thus, can be tested for other requests to verify their probiotic efficiency.
Similar antibiotic profil of the selected LAB were observed in several studies, and it was shown that the Lactobacilli are resistant to some antibiotics like: Nalidixic acid, streptomycin, vancomycin and gentamycin.12,16,19,27,32 However, Argyri, et al.27 reported that the resistance of Lactobacillus genus to aminoglycoside antibiotics is considered as intrinsic. On the other hand, Peres, et al.19 and Kumar and Kumar16 suggested that the resistance of probiotics to antibiotics can be an advantageous to survive in the gastrointestinal tract during antibiotic treatment.
The LAB have a big interest in terms of food safety and prevention of intestinal infections6; through, their production of anti-microbial agent, and which is considered as one criterion for the probiotic selection.16,33 So, in this study, inhibitive potential of selected LAB was determined, and it revealed that strains have good capacity to inhibit some pathogens like: Pseudomonas sp., Staphylococcus aureus and E. coli. In fact, several antagonistic compounds secreted by LAB including: organic acids, especially lactic acid, hydrogen peroxide and bacteriocins, which inhibit pathogens in the gastrointestinal tract and preserve animal or human foods , have been mentioned in the literature.33,34
Adhesion to the epithelial cells is considered, also, as an important selection criterion for a probiotic organism1. In this study, adhesion of some selected strains to epithelial cells was shown without fixation step, which gives evidence of the natural adhesion of the selected bacteria to the chicken epithelial cells. The adhesion ability of LAB to epithelial cells was observed in many studies.10,11,26,27,29 Also, the biochemical characterization of the adhesion suggested that glycoproteins and heat-labile carbohydrates are involved in the adherence mechanism11, and it was concluded by Mayra Maukinen, et al.35 and by Fernandez, et al.11 that the adhesion ability of Lactobacilli is strain-dependent.
This study confirms that the Algerian fermented products could constitute an interesting source to obtain Lactobacillus spp. strains with probiotic potential, such as, L. plantarum and L. fermentum. Results show that four isolates among the fourteen: Lactobacillus plantarum OV13, Lactobacillus sp. OV15, Lactobacillus fermentum E161 and Lactobacillus sp. E631, have the best probiotic performance, due to their high resistance to bile and to simulated gastric conditions, their antibiotic resistance, their antibacterial activity and also their epithelial cells adhesion. Therefore, the potential effect of these strains on the animal’s food should be investigated for the purpose of its use as starter cultures for animal’s food conservation (silage) and thus, use it as a reliable food vehicle into the animal gastrointestinal tract, and finally, the follow-up of their effect on the host health.
ACKNOWLEDGMENTS
Our thanks go to Pr. Thierry BERGES, Poitiers University, for his assistance in the molecular identification, and also to Mr Toufik SAHRAOUI, LMESM Laboratory, USTO University, for SEM analysis.
- Van Tassell, M.L., Miller, M.J. Lactobacillus adhesion to mucus. Nutrients,2011; 3(5):613-636.
- Metchnikoff, E. The prolongation of life; optimistic studies. New York & London : G.P. Putnam’s Sons, 1908.pp 380
- Kalsum, U., Soetanto, H., Sjofjan, O. Influence of a probiotic containing lactobacillus fermentum on the laying performance and egg quality of Japanese quails. Int. J. Poult. Sci.,2012; 11(4):311-315.
- Perdigon, G., Fuller, R., Raya, R. Lactic acid bacteria and their effect on the immune system. Curr. Issues Intestinal Microbiol.,2001; 2(1):27-42.
- Nicolas, J.L., Gatesoupe, J., Frouel, S., Bachere, E., Gueguen, Y. Quelles stratégies alternatives aux antibiotiques en aquaculture? INRA Production animale,2007; 20(3):253-258.
- Herich, R., Levkut, M. Lactic acid bacteria, probiotics and immune system. Vet. Med- Czech. ,2002; 47(6):169-180.
- Uccello, M., Malaguarnera, G., Basile, F., D’agata, V., Malaguarnera, M., Bertino, G., Vacante, M., Drago, F., Biondi, A. Potential role of probiotics on colorectal cancer prevention. BMC surgery,2012; 12(1):1-8.
- Davoodabadi, A., Dallal, M.M.S., Foroushani, A.R., Douraghi, M., Harati, F.A. Antibacterial activity of Lactobacillus spp. isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe,2015; 34:53-58.
- Pan, X., Chen, F., Wu, T., Tang, H., Zhao, Z. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Control,2009; 20(6):598-602.
- Jamaly, N., Benjouad, A., Bouksaim, M. Probiotic potential of Lactobacillus strains isolated from known popular traditional moroccan dairy products. Br. Microbiol. Res. J.,2011; 1(4):79-94.
- Fernandez, M., Boris, S., Barbes, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol.,2003; 94(3):449-455.
- Cebeci, A., Gurakan, C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol.,2003; 20(5):511-518.
- Thirabunyanon, M., Boonprasom, P., Niamsup, P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol. Lett.,2009; 31(4):571-576.
- Kacem, M., Karam, N.E. In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. Int. J. Probiotics Prebiotics,2006; 1(1):27-32.
- Saelim, K., Sohsomboon, N., Kaewsuwan, S., Maneerat, S. Probiotic properties of Enterococcus faecium CE5-1 producing a bacteriocin-like substance and its antagonistic effect against antibiotic-resistant enterococci in vitro. Czech. J. Anim. Sci.,2012; 57(11):529-539.
- Kumar, A., Kumar, D. Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobe,2015; 33:117-123.
- Ocana, V.S., Nader-Macias, M.E. Adhesion ability of Lactobacillus to vaginal epithelial cells. Public Health Microbiology: Methods and Protocols,2004:441-445.
- Ruiz, L., Margolles, A., Sanchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol.,2013; 4:396.
- Peres, C.M., Alves, M., Hernandez-Mendoza, A., Moreira, L., Silva, S., Bronze, M.R., Vilas-Boas, L., Peres, C., Malcata, F.X. Novel isolates of lactobacilli from fermented Portuguese olive as potential probiotics. LWT-Food. Sci. Technol.,2014; 59(1):234-246.
- Khedid, K., Faid, M., Mokhtari, A., Soulaymani, A., Zinedine, A. Characterization of lactic acid bacteria isolated from the one humped camel milk produced in Morocco. Microbiol. Res.,2009; 164(1):81-91.
- Mahmoudi, I., Moussa, O.B., Khaldi, T.E.M., Kebouchi, M., Soligot, C., Le Roux, Y., Hassouna, M. Functional in vitro screening of Lactobacillus strains isolated from Tunisian camel raw milk toward their selection as probiotic. Small Ruminant Res.,2016; 137:91-98.
- Kacem, M., Karam, N.E. Microbiological study of naturally fermented Algerian green olives: isolation and identification of lactic acid bacteria and yeasts along with the effects of brine solutions obtained at the end of olive fermentation on Lactobacillus plantarum. Grasas Aceites,2006; 57(3):292-300.
- Hurtado, A., Reguant, C., Bordons, A., Rozes, N. Lactic acid bacteria from fermented table olives. Food Microbiol.,2012; 31(1):1-8.
- Yang, J., Cao, Y., Cai, Y., Terada, F. Natural populations of lactic acid bacteria isolated from vegetable residues and silage fermentation. J. Dairy Sci.,2010; 93(7):3136-3145.
- Pang, H., Qin, G., Tan, Z., Li, Z., Wang, Y., Cai, Y. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol.,2011; 34(3):235-241.
- Gu, R.X., Yang, Z.Q., Li, Z.H., Chen, S.L., Luo, Z.L. Probiotic properties of lactic acid bacteria isolated from stool samples of longevous people in regions of Hotan, Xinjiang and Bama, Guangxi, China. Anaerobe,2008; 14(6):313-317.
- Argyri, A.A., Zoumpopoulou, G., Karatzas, K.A.G., Tsakalidou, E., Nychas, G.J.E., Panagou, E.Z., Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol.,2013; 33(2):282-291.
- Dunne, C., O’Mahony, L., Murphy, L., Thornton, G., Morrissey, D., O’Halloran, S., Feeney, M., Flynn, S., Fitzgerald, G., Daly, C. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr.,2001; 73(2):386-392.
- Lahteinen, T., Malinen, E., Koort, J.M., Mertaniemi-Hannus, U., Hankimo, T., Karikoski, N., Pakkanen, S., Laine, H., Sillanpaa, H., Soderholm, H. Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe,2010; 16(3):293-300.
- Lin, W.H., Yu, B., Jang, S.H., Tsen, H.Y. Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe,2007; 13(3):107-113.
- Balcazar, J.L., Vendrell, D., de Blas, I., Ruiz-Zarzuela, I., Muzquiz, J.L., Girones, O. Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture,2008; 278(1):188-191.
- Florez, A.B., Delgado, S., Mayo, B. Antimicrobial susceptibility of lactic acid bacteria isolated from a cheese environment. Can. J. Microbiol.,2005; 51(1):51-58.
- Settanni, L., Corsetti, A. Application of bacteriocins in vegetable food biopreservation. Int. J. Food Microbiol.,2008; 121(2):123-138.
- Caplice, E., Fitzgerald, G.F. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol.,1999; 50(1):131-149.
- Mayra Maukinen, A., Mnninen, M., Gyllenberg, H. The adherence of lactic acid bacteria to the columnar epithelial cells of pigs and calves. J. Appl. Bacteriol.,1983; 55(2):241-245.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


