ISSN: 0973-7510
E-ISSN: 2581-690X
Effective rhizosphere colonization by plant growth promoting rhizobacteria (PGPR) is a prerequisite for its persistence performance under plant-root soil system. Therefore, to obtain effective strains, we have evaluated twelve strains of fluorescent Pseudomonas sp. from wheat rhizosphere exhibiting multifarious plant growth promoting (PGP) activities. These strains formed varying level of biofilm in vitro on abiotic surface. Two strains MW3 and MW4 with promising PGP activity but differing in biofilm forming ability. The biofilms were characterized by crystal violet assay, scanning electron and confocal laser scanning microscopy. The MW4 strain, a strong biofilm former is identified by16S rRNA gene sequence analysis. Strain MW4 showed strong biofilms compared to MW3. Similarly, wheat seeds treated with MW3 and MW4 revealed enhanced rhizospheric colonization by MW4 strain significantly (P ≤ 0.05) in higher number (logCFU g-1) up to 45 days after sowing compared to MW3. These findings suggested that strong biofilm forming ability of MW4 might have resulted in enhanced rhizosphere colonization. Therefore, biofilm forming ability of PGPR should be considered as an additional criterion in screening of fluorescent Pseudomonas sp.
Biofilm; Pseudomonas fluorescens; Triticum aestivum; root colonization; 16S rRNA gene; confocal laser scanning microscopy.
Rhizobacteria promoting plant growths are diverse and contribute in crop productivity by their various direct and indirect PGP activities1. Fluorescent Pseudomonads have been found as one of the important PGPRs exploited in crop production2. However, the performance of microbial strains used as biofertilizer is not always consistent due to several biotic and abiotic factors3. Therefore, effective rhizosphere colonization by microbial inoculants under field conditions is most critical to promote plant growth consistently. It is now commonly accepted that under natural environment bacteria exist mainly in biofilm mode rather than planktonic state. Biofilms are defined as highly organized microbial communities encased in self-produced extracellular matrix4. It is believed that bacterial strains exhibiting biofilm associated characteristics such as flagellar motility, type IV pili, hydrophobicity and production of exopolysaccharides might be helpful in effective biofilm formation and rhizosphere colonization5-8. Therefore, in this study, fluorescent Pseudomonas strains exhibiting multiple plant growth promoting traits were tested for their biofilm formation. Most effective biofilm forming strain (MW4) was further compared with moderate biofilm forming fluorescent Pseudomonas (MW3) to assess possible relationship between biofilm development and wheat rhizosphere colonization.
Isolation of fluorescent Pseudomonas
Wheat plants cultivated locally in the vicinity of Aligarh city, India were uprooted carefully and rhizospheric soil was collected. Briefly, soil tightly adhered to roots were collected by vortexing the roots in 0.85% NaCl solution to prepare stock suspension. Hundred microlitre of appropriately diluted sample was spread on King’s B agar plates and incubated for 48 h at 28°C. Typical colonies producing yellow-green fluorescence detected under UV light were selected and purified. The cultures were given a strain number and maintained in the laboratory by subculturing. One set of culture was preserved at -80°C in glycerol broth. The fluorescent Pseudomonas strains were biochemically characterized and tentatively identified using standard methods9-10.
Assay for production of plant growth promoting activities
Plant growth promoting activities of isolated bacteria was determined using standard methods as described previously6 with little modifications. A brief description is given here. Production of Indole-3-acetic acid (IAA) was analyzed using the standard method of Loper and Scroth11 with the amendment of media by tryptophan (100 µg mL-1). Siderophore production was studied by growing the cultures in Modi medium. Catechol-type phenolates were measured as described by Reeves et al.12. Phosphate solubilization was studied by using standard method13.
The exopolysaccharides (EPS) production was done by the method of Mody et al14. 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity of the strains was checked by growing them onto DF (Dworkin and Foster’s) salts minimal agar medium15 amended with 3.0 mM ACC (Sigma-Aldrich, USA) instead of (NH4)2SO4 as nitrogen source16. Bacterial strains were also evaluated for their antifungal activity against Fusarium oxysporum (MTCC-3930), Fusarium ciceri (ITCC-3636), Fusarium solani (ITCC- 6953), Aspergillus niger (ITCC-6370), Alernaria alternata (ITCC-6778) by dual culture assay17.
Assay for biofilm development
The biofilm formation was determined in 96 well polystyrene microtitre plates as described by O’Toole and Kolter18. Briefly, bacteria were grown in the tryptic soy broth (TSB, HiMedia, India) at 28 °C for 24 h. Each well was filled with 200 µL (≈107 CFU mL-1) of culture in fresh medium and allowed to grow for 24 h at 28 °C. The wells were washed with phosphate buffer saline (PBS), followed by staining with 0.1% crystal violet for 20 min. Unbound strain was removed by washing each well with PBS. In each well ethanol (95%) was added to solubilize the bound dye and absorbance was recorded at 590 nm using micro plate reader (ThermoScientific Multiskan EX, UK). Based on absorbance value of dye, the strains were distinguished as strong and moderate biofilm formers according to the criteria adopted by Christensen et al19.
Identification of MW4 strain based on 16S rRNA gene sequence analysis
The methods adopted at Macrogen (South Korea) for molecular characterization is briefly described below. Genomic DNA was extracted from MW4 and used as template in a 30 µL reaction mixture by using an EF-TaqDNA polymerase (SolGent, South Korea). The primers 518F (5’-CCAGCAGCCGCGGTAATACG-3´) and 800R (5´-TACCAGGGTATCTAAT CC-3´) were used for the PCR (Polymerase chain reaction). The PCR reaction was performed with 20 ng of genomic DNA as the template Activation of Taq polymerase at 95°C for 2 min, 35 cycles of 95°C for 1 min, 55°C, and 72°C for 1 min each were performed, finishing with a 10 min step at 72°C. The amplified PCR products were purified with a multi screen filter plate (Millipore Corp., USA). Sequencing reaction was performed using a PRISM BigDye Terminator v3.1 Cycle Sequencing Kit. The DNA samples containing the extension products were added to Hi-Di formamide (Applied Biosystems, CA, USA). The mixture was incubated at 95°C for 5 min, followed by 5 min on ice and thenanalyzed by ABI Prism 3730XL DNA analyzer (Applied Biosystems, Macrogen Inc, Seoul, South Korea). The resulting nucleotide sequence measuring 997 bp in size was submitted to GenBank sequence database (accession number KF555376). The sequence obtained were analysed using Basic Local Alignment Search Tool (BLASTN) and matched against known bacterial sequences present in NCBI GenBank data base (http: /www.ncbi.nlm.nih.gov). Related sequences obtained from the database were used to construct a phylogenetic tree using MEGA5 software.
Rhizosphere colonization assay
For colonization study under non sterile soil condition, rifampicin resistant mutant of the two parent strains MW4 and MW3 was selected based on sequential exposure of rifampicin and selecting the mutant on rifampicin amended Kings B agar plate and checked their stability for several generations. Triticum aestivum L. (var. PBW-343) surface sterilized seeds were treated with rifampicin resistant mutants MW3 and MW4 (≈ 2.8×107 CFU seed-1) separately and sown in natural loam soil. Collection of rhizosphere and rhizoplane samples (five plants) was made at 15, 30, and 45 days after sowing. Serial dilutions from rhizosphere and rhizoplane samples were prepared separately in normal saline solution (NSS) and plated onto King’s B agar medium containing 800µg mL-1 rifampicin. Untreated plants maintained in similar condition were also subjected to enumerate presence of naturally occurring rifampicin resistant bacteria capable of growing on King’s B agar medium at 800µg mL-1 rifampicin. Only fluorescent colonies grown on rifampicin amended plates were counted.
Biofilm formation on glass cover slips by MW3 and MW4
Crystal violet assay with little modifications as described by O’Toole and Kolter18 was performed to establish the biofilm development on glass coverslips. Briefly, bacterial strains were grown in tryptic soy broth (TSB) as described earlier. A 12 well tissue culture plate with flat bottom containing sterile coverslips were filled with TSB and 100 µL of inoculum (≈107 CFU mL-1) per well and incubated statically at 28 °C for 48 h. After growth coverslips were removed and rinsed in PBS followed by staining with 0.1% crystal violet for 20 min. After staining, coverslips were washed with PBS to remove excess dye and examined under a light microscope (Olympus BX60, Japan) at 100X.
Confocal laser scanning microscopy of biofilms by MW3 and MW4
The biofilms formed on coverslips as mentioned above were stained with acridine orange (Sigma-Aldrich, USA). Microscopic observations and image acquisitions were performed using confocal laser scanning microscopy (CLSM) (Olympus Fluoview FV1000) using standard protocol.
Scanning electron microscopy (SEM) and CLSM and of biofilms formed on the roots of T. aestivum
Triticum aestivum L. (var. PBW 343) surface sterilized seeds were sown in sterile sand-soil mixture for raising seedlings. After 15 days of growth, the plant was uprooted, and roots were washed with sterile distilled water and treated with bacterial culture by dipping the roots in 10 mL diluted culture of fluorescent Pseudomonas MW4 and MW3 (≈105 CFU mL-1) prepared in TSB. In the control plants, sterile distilled water (SDW) was used instead of culture. After 24 hours the root samples was fixed in 2.5% glutaraldehyde and washed in the same buffer followed by fixation in osmium tetraoxide. The root were dehydrated by treating with graded concentrations of ethanol and dried in CO2 with a critical point-dryer, mounted on stubs, and shadowed with gold, (Sputter Coater-Polaron SC7640 UK) for viewing under an scanning electron microscopy (SEM) (Carl ZeissEVO 40, Germany). For CLSM, 24 h-treated root samples were first stained with acridine orange and images were acquired as mentioned above.
Isolation, characterization and screening of fluorescent Pseudomonas
After initial screening of 56 strains suspected to be fluorescent Pseudomonas, twelve strains were selected based on their multiple PGP traits (≥ 3), where MW4 exhibited a comparatively higher level of activity for phosphate solubilization, indole acetic acid, and siderophore production (Fig. 1a) along with antifungal activity against Aspergillus niger, Fusarium oxysporum, and Fusarium ciceri. Further, these 12 strains were tested for biofilm forming ability in vitro on polystyrene microtiter plate. The data obtained indicates variations in their biofilm forming ability. The strains were grouped into moderate biofilm former (MW3, MW5, MW6, MW8, MW10, and MW11) and strong biofilm former (MW1, MW2, MW4, MW7, MW9, and MW12) (Fig. 1b).
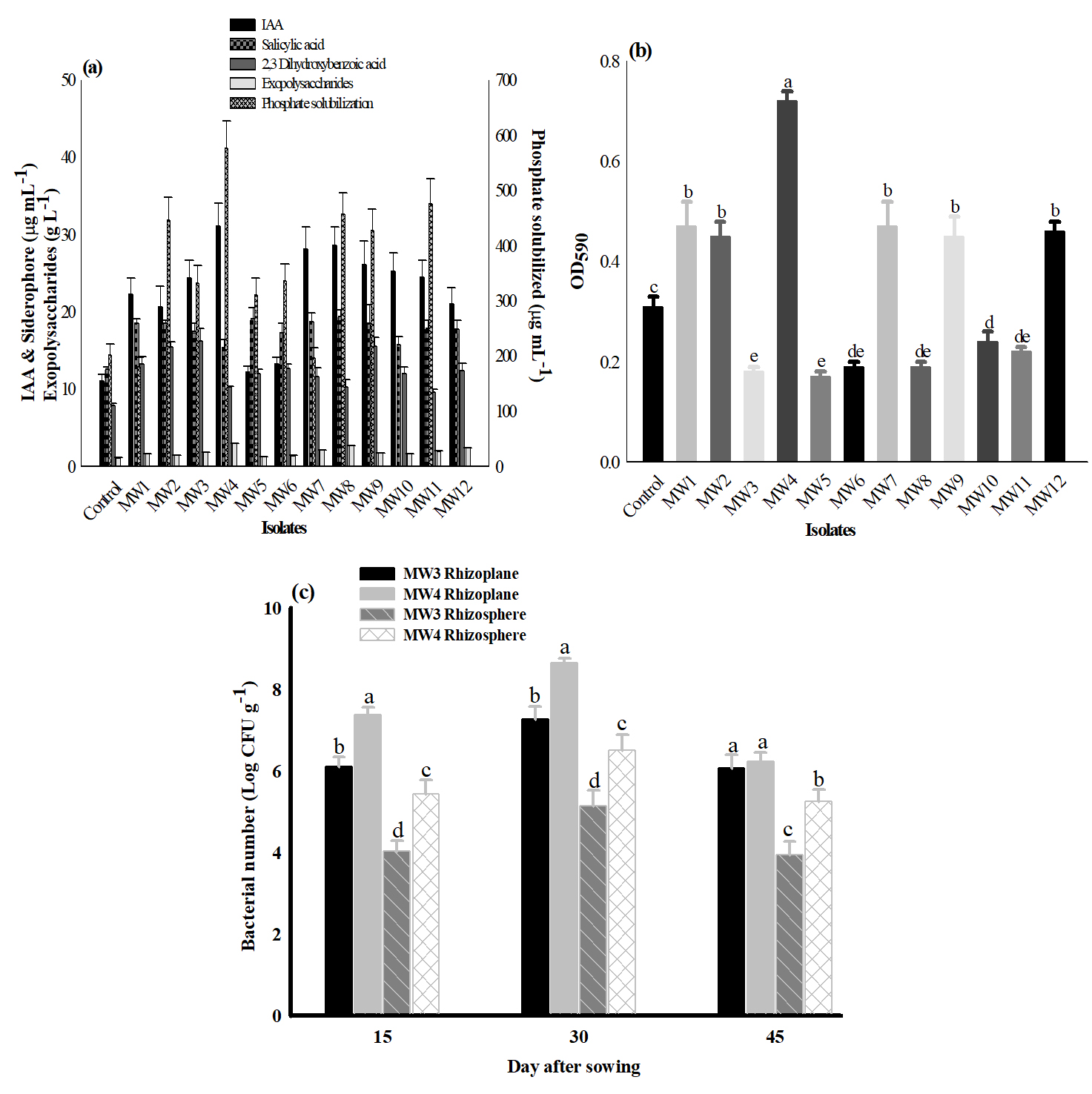 Fig. 1. Plant growth promoting activities and biofilm formation by fluorescent Pseudomonas strains. (a) Plant growth promoting traits of test strains. (b) Biofilm formation on 96 well microtiter plates under stationary conditions. Bar values followed by a different letters are significantly different (a = 0.05) in Duncan’s multiple range test. MTCC-1749 was used as positive control. (c)Survival and colonization of MW3, MW4 (rifampicin resistant mutant) with wheat rhizoplane and rhizosphere in natural soil conditions. Bar values followed by a different letters are significantly different (a = 0.05) in Duncan’s multiple range test.
Fig. 1. Plant growth promoting activities and biofilm formation by fluorescent Pseudomonas strains. (a) Plant growth promoting traits of test strains. (b) Biofilm formation on 96 well microtiter plates under stationary conditions. Bar values followed by a different letters are significantly different (a = 0.05) in Duncan’s multiple range test. MTCC-1749 was used as positive control. (c)Survival and colonization of MW3, MW4 (rifampicin resistant mutant) with wheat rhizoplane and rhizosphere in natural soil conditions. Bar values followed by a different letters are significantly different (a = 0.05) in Duncan’s multiple range test. Biofilm studies on Pseudomonas fluorescens MW4 and MW3 strains in vitro
On the basis of biofilm forming capability in vitro, plant growth promoting traits and effect on seed germination and seedling growth, two strains (MW4 and MW3) were selected for further study. Biofilm formation by P. fluorescens MW4 and MW3 were further performed on glass coverslips (Fig. 2a). The results demonstrated that strain MW4 developed dense and thick biofilms. Further, biofilm was analyzed by CLSM (Fig. 2b), which showed bacterial aggregation and formation of biofilm by MW4 whereas less surface adhered bacteria were observed in MW3 (moderate biofilm former). The average thickness of the MW4 biofilm was found to be 19.29µm as evident from Z-stack analysis (Fig. 2c).
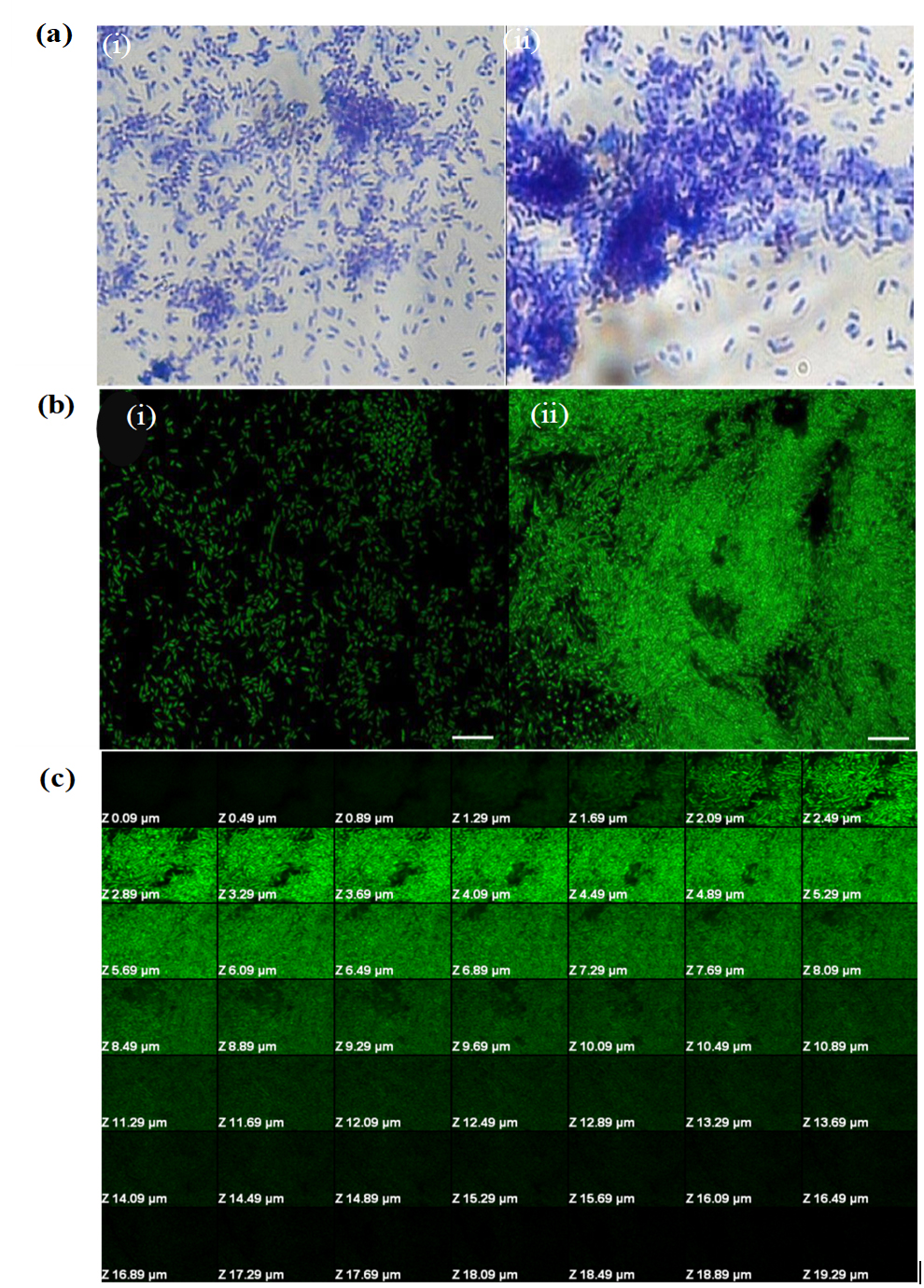 Fig. 2. (a) Biofilm formation on glass coverslips by fluorescent Pseudomonas strains using crystal violet assay. (i) MW3,(ii) MW4.(b) CLSM images of biofilm formed by fluorescent Pseudomonas strains on glass coverslip under stationary conditions. (i)MW3,(ii) MW4.Biofilms were stained with acridine orange. Magnification is 100x. Scale bars = 5 µm. (c) Confocal laser scanning micrographs of Z-stack analysis of Pseudomonas fluorescens (MW4) biofilm formed on glass coverslip.
Fig. 2. (a) Biofilm formation on glass coverslips by fluorescent Pseudomonas strains using crystal violet assay. (i) MW3,(ii) MW4.(b) CLSM images of biofilm formed by fluorescent Pseudomonas strains on glass coverslip under stationary conditions. (i)MW3,(ii) MW4.Biofilms were stained with acridine orange. Magnification is 100x. Scale bars = 5 µm. (c) Confocal laser scanning micrographs of Z-stack analysis of Pseudomonas fluorescens (MW4) biofilm formed on glass coverslip. Root colonization by strong and moderate biofilm forming bacterial strains
Root colonization was performed by selected two strains MW4 and MW3. The Pseudomonas fluorescens MW4 treated seeds yielded log 8.44 CFU g-1 seed. The rhizospheric and rhizoplane viable count of the strain was carried out at 15, 30 and 45 days after sowing and change in bacterial population was observed (Fig. 1c). In rhizoplane, the population of MW4 decreased one log after 15 days of sowing (log 7.38 CFU g-1 root wt) compared to MW3 (log 6.11 CFU g-1 root wt). There was an increase of CFU count after 30 days of sowing but decrease after 45 days. Whereas, in rhizosphere the mean values of viable counts for MW4 and MW3 were log 5.44 and 4.04 CFU g-1 of rhizosphere soil respectively at 15 days after sowing. At 30 days, a little increase in the population of both strains was recorded but a declining trend was observed after 45 days of sowing. Strain MW4 colonized both rhizoplane and rhizosphere more effectively as compared to MW3 even up to 45 days after sowing.
Biofilm studies of MW4 and MW3 in vivo
The behavior of the MW4 and MW3 strains during their interactions with wheat seedlings was evaluated using SEM and CLSM. Root sections of wheat bacterized with MW4 and MW3 were examined. The results revealed that cells of MW4 were consistently distributed on the surface of roots (Fig. 3d-j), whereas MW3 showed decreased root colonization and biofilm formation (Fig. 3a-c). A biofilm consisting of bacterial cells and a net-like material suggest extracellular matrix formation (Fig. 3h-j). Less adherent microbial cells were observed in the negative control (untreated) on any of the observed root samples (Fig. 3k-n). Similar pattern of biofilm on root surface was revealed for MW4 when analyzed with CLSM (Fig. 4c-d) compared to MW3 which showed moderate biofilm formation (Fig. 4a-b). The average thickness of the MW4 biofilm formed on wheat root surface was found to be 24.92 µm as evident from Z-stack analysis (Fig. 4e). The strain was found a strong biofilm former as well as colonized roots by dense biofilm development compared to moderate biofilm former MW3.
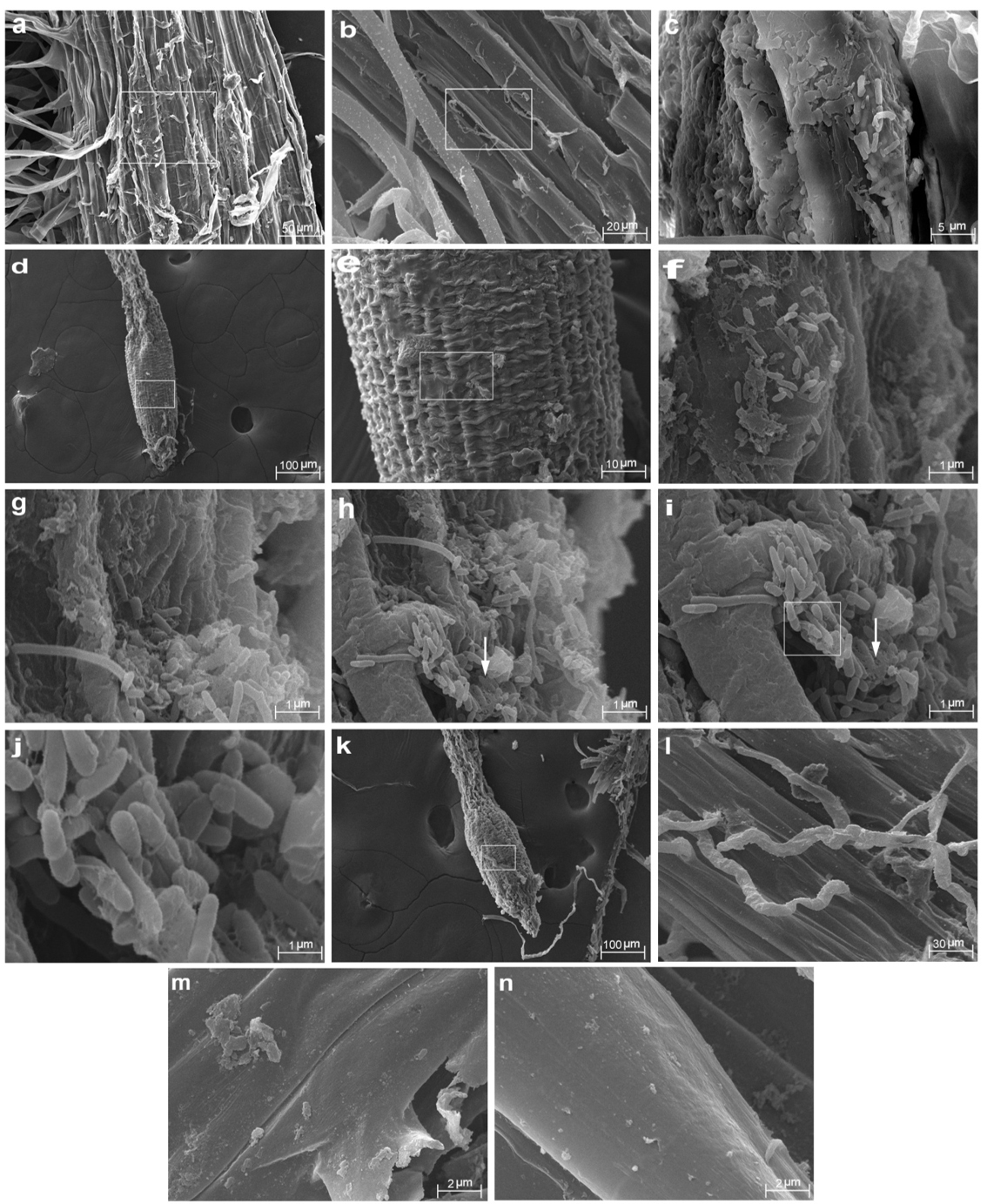 Fig. 3. Scanning electron micrographs of Triticum aestivum seedling roots colonized by Pseudomonas fluorescens MW3 and MW4. (a) Colonization and biofilm formation on the surface of the root zone by MW3 (moderate biofilm former). (b) Magnified image of the framed region shown in figure 3a. (c) Colonization and biofilm formation of the framed region shown in figure 3b. (d) Colonization and biofilm formation on Triticum aestivum roots by MW4. (e) Magnified image of the framed region shown in figure 3d.(f) Large scale colonization of the framed region shown in figure 3e. (g) Biofilm formation by Pseudomonas fluorescens MW4. (h-i) Extracellular matrix production by biofilm forming MW4 (white arrow). (j) Magnified image of the framed region shown in figure 3i. (k-n) SEM image of root zone of control (untreated) seedling. (l) Magnified image of the framed region shown in figure 3k.
Fig. 3. Scanning electron micrographs of Triticum aestivum seedling roots colonized by Pseudomonas fluorescens MW3 and MW4. (a) Colonization and biofilm formation on the surface of the root zone by MW3 (moderate biofilm former). (b) Magnified image of the framed region shown in figure 3a. (c) Colonization and biofilm formation of the framed region shown in figure 3b. (d) Colonization and biofilm formation on Triticum aestivum roots by MW4. (e) Magnified image of the framed region shown in figure 3d.(f) Large scale colonization of the framed region shown in figure 3e. (g) Biofilm formation by Pseudomonas fluorescens MW4. (h-i) Extracellular matrix production by biofilm forming MW4 (white arrow). (j) Magnified image of the framed region shown in figure 3i. (k-n) SEM image of root zone of control (untreated) seedling. (l) Magnified image of the framed region shown in figure 3k.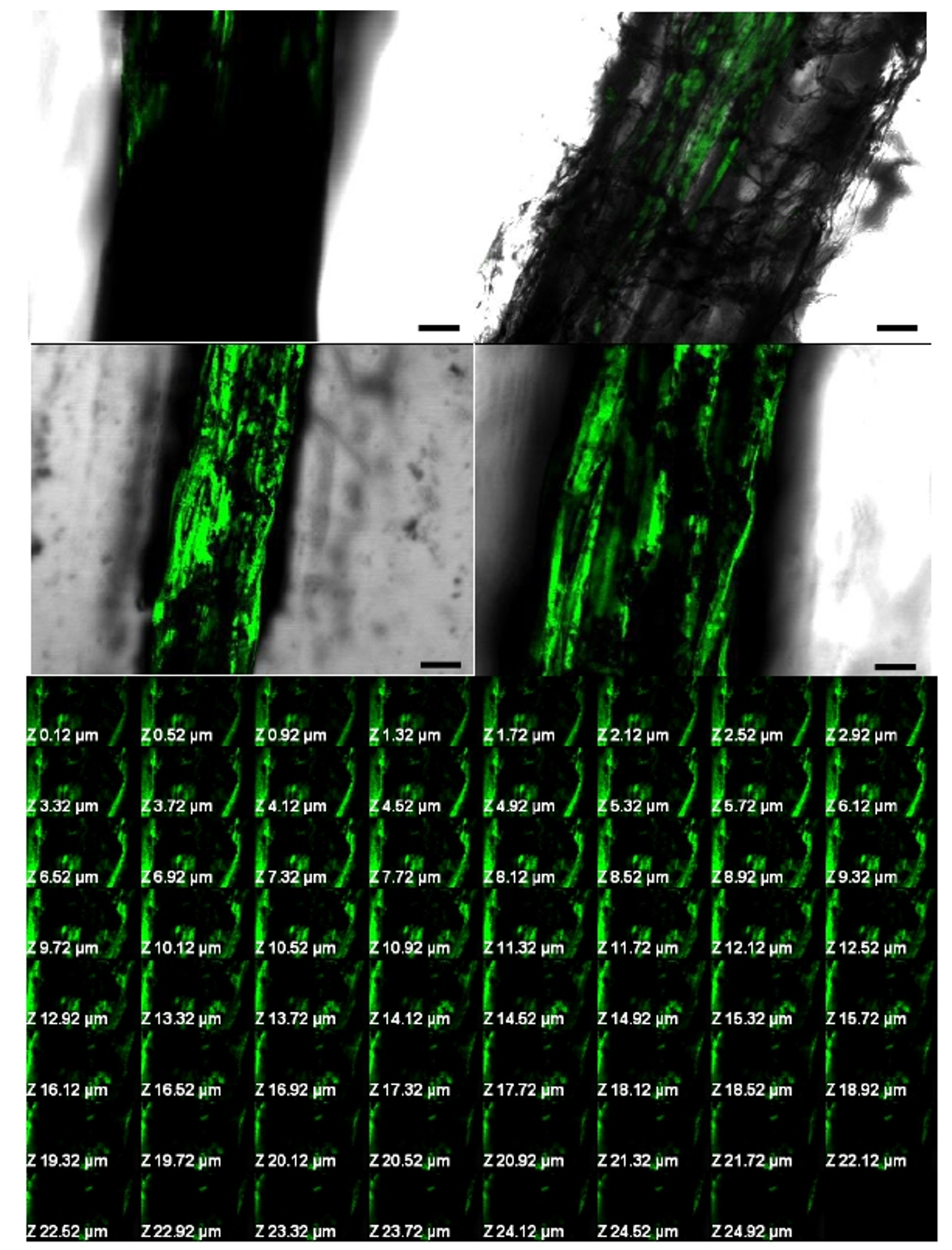 Fig. 4. Confocal microscopy images of MW3 and MW4 biofilm grown on Triticum aestivum root surface. (a) Colonization and biofilm formation by MW3 (moderate biofilm former). (b) Longitudinal section of primary root showing the aggregation of cells as biofilm formed by MW3. (c) Longitudinal section of primary root showing aggregation, colonization and strong biofilm formation on the root surface by MW4 (strong biofilm former). (d) Magnified image of figure 4c. Biofilms were stained with acridine orange. Magnification is 20x. Scale bars = 50 µm. (e) Confocal laser scanning micrographs of Z-stack analysis of Pseudomonas fluorescens (MW4) biofilm formed on Triticum aestivum seedling roots after 15 days of growth.
Fig. 4. Confocal microscopy images of MW3 and MW4 biofilm grown on Triticum aestivum root surface. (a) Colonization and biofilm formation by MW3 (moderate biofilm former). (b) Longitudinal section of primary root showing the aggregation of cells as biofilm formed by MW3. (c) Longitudinal section of primary root showing aggregation, colonization and strong biofilm formation on the root surface by MW4 (strong biofilm former). (d) Magnified image of figure 4c. Biofilms were stained with acridine orange. Magnification is 20x. Scale bars = 50 µm. (e) Confocal laser scanning micrographs of Z-stack analysis of Pseudomonas fluorescens (MW4) biofilm formed on Triticum aestivum seedling roots after 15 days of growth.Biofilm forming rhizobacteria are expected to colonize plant roots more stably and can remain active more consistently. To test this hypothesis, fluorescent Pseudomonas strains isolated from wheat (Triticum aestivum L.) rhizosphere and exhibiting multiple PGP activities and biofilm forming ability in vitro were studied for their potential for rhizosphere colonization through biofilm formation.
These strains were tested and identified as Pseudomonas sp. and strain MW4 was identified as Pseudomonas fluorescens by molecular techniques. The strain MW4 was further selected due to strong biofilm formation ability and increased level of PGP activities.
PGPR including fluorescent Pseudomonas are known to exert plant growth promoting properties by both direct mechanisms (assisting nutrient acquirement, adjusting phytohormone concentration, nitrogen fixation, phosphate solubilization and sequestering iron) and indirect mechanisms such as production of antibiotics, lytic enzymes and siderophores, competition, induced systemic resistance1. Pseudomonas fluorescens MW4 produced auxins, solubilized phosphate, siderophore and ACC deaminase activity. Presence of multiple PGPR traits observed in this study is in agreement with the report of Zabihi et al20. Significant production of exopolysaccharides by PGPR is expected to confer protection against the strained conditions and assist in bacterial root colonization21. Moreover, exopolysaccharides have water retaining and adhesive characteristics and play a significant role in stabilization of soil aggregates and the regulation of nutrients and water flow across plant roots22. Strain MW4 was assessed for its potential to form biofilm on roots of the wheat plant under natural aseptic conditions which is evident from SEM and CLSM. The strain MW4 colonized plant root and form biofilm, indicating that such activity might be helpful in protecting the plant from root borne pathogens23. To evaluate this, we perform an experiment for their survival in rhizoplane and rhizosphere. The findings have clearly demonstrated better rhizosphere colonization by strong biofilm former MW4 compared to moderate biofilm former, MW3 under natural condition.
Based on above findings it can be concluded that Pseudomonas fluorescens MW4 is a strong biofilm former and competent root colonizing PGPR which could be developed as inoculant to assess its performance under field condition. Further mutants of these cultures deficient in biofilm formation need to be tested to explore the role of biofilm and other traits in improving the rhizosphere ecological fitness of the bacteria.
Acknowledgements
The author MMA is thankful to UGC, New Delhi for providing fellowship. We are also grateful to AIRF, JNU, New Delhi, for SEM and CLSM observation.
Conflict of interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
None
Data availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Glick BR. 2012. Plant growth-promoting bacteria: mechanism and application, pp.1-15.Scientifica.
Crossref - Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moenne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol, 2009; 48: 505-512.
Crossref - RaaijmakersJM. 2015. The minimal rhizosphere microbiome, pp. 411-417. In Lugtenberg, B., (ed.) Principles of Plant-Microbe Interactions. Springer: Heidelberg.
Crossref - Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol, 2004; 2: 95–108.
Crossref - Velmourougane K, Prasanna R, Saxena AK. Agriculturally important microbial biofilms: Present status and future prospects. J Basic Microbiol, 2017; 57(7):548-573.
Crossref - Altaf MM, Ahmad I. Plant Growth Promoting Activities, Biofilm Formation and Root Colonization by Bacillus sp. isolated from Rhizospheric Soils. J Pure Appl Microbiol, 2016; 10 (1):109-120.
- Altaf MM., Ahmad I. In vitro and In vivo biofilm formation by Azotobacter strains and its relevance to rhizosphere colonization. Rhizosphere, 2017; 3: 138–142.
Crossref - Ansari FA, Ahmad I. Fluorescentpseudomonas -FAP2 and Bacillus licheniformis interact positively in biofilm mode enhancing plant growthand photosynthetic attributes. Sci Rep, 2019; 9(1):4547.
Crossref - Cappuccino JC, Sherman N, 2001. Microbiology: A Laboratory Manual. Benjamin/ Cummings: New York.
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST, 1994. Bergy’s Manual of Determinative Bacteriology, ninth ed. Williams and Wilkins Pub: MD, USA.
- Loper JE, Scroth MN. Influence of bacterial sources on indole-3 acetic acid on rootelongation of sugar beet. Phytopathol,1986; 76:386-389.
Crossref - Reeves MW, Pine L., Neilands JB, Balows A. Absence of siderophore activity in Legionella species grown in iron-deficient media. J Bacteriol, 1983; 154: 324-329.
- King JE, 1932. The colorimetric determination of phosphorus. BiochemJ, 1932; 26: 292-297.
Crossref - Mody BR, Bindra MO, Modi VV. Extracellular polysaccharides of cowpea rhizobia: compositional and functional studies. Arch Microbiol, 1989; 1: 2-5.
Crossref - Dworkin M, Foster J. Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol, 1958; 75: 592–601.
- Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant, 2003; 118: 10-15.
Crossref - Landa BB, Hervאs A, Bettiol W, Jimיnez-Dמaz RM. Antagonistic activity of bacteria from the chickpea rhizosphere against Fusarium oxysporumf. sp. ciceris. Phytoparasitica, 1997; 25:305-318.
Crossref - O’Toole GA, Kolter R. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol, 1998; 28: 449-461.
Crossref - Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol, 1985; 22: 996-1006.
- Zabihi HR, Savaghebi GR, Khavazi K, Ganjali A, Miransari M. Pseudomonas bacteria and phosphorous fertilization, affecting wheat (Triticum aestivumL.) yield and P uptake under greenhouse and field conditions. ActaPhysiol Plant,2011; 33: 145-152.
Crossref - Bogino PC, Oliva MM, Sorroche FG, Giordano W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int J Mol Sci, 2013; 14:15838–15859.
Crossref - Roberson EB, Firestone MK. Relationship between desiccation and exopolysaccharide production in soil Pseudomonas sp. Appl Environ Microbiol, 1992; 581284-1291.
- Timmusk S, Behers L, Muthoni J, Muraya A, Aronsson A-C. Perspectives and challenges of microbial application for crop improvement. Front Plant Sci, 2017; 8:49.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


