ISSN: 0973-7510
E-ISSN: 2581-690X
The COVID-19 caused by a new type of coronavirus has emerged from China and led to thousands of death globally. Despite the efforts engaged in studying this newly emerged virus and searching for its treatment, the understanding of the COVID-19 drug and target protein interactions still represent a key challenge. Several molecules have demonstrated In-Vitro activity against the SARS-CoV-2 virus and/or potential clinical benefit in observational and non-randomized studies. Randomized clinical trials of an appropriate size are currently ongoing to establish the efficacy of these therapeutic proposals. Herein, concerning these diverse guidelines and therapeutic suggestions of different approaches to the treatment, this research aims to provide a molecular analysis of the interaction between the principal molecules cited in bibliography and the active protease site of the virus.
COVID-19, Pharmacological treatment, Protease, docking, SARS-CoV2
In order to face the SARS-CoV-2 pandemic, several therapeutic protocols have been issued across the world, these proposals are based on the repositioning of drugs or on clinical trials being conducted in China as the country where this pandemic first emerged. However, given the lack of molecules that can selectively inhibit the virus, the use of this methodology remains the appropriate temporary solution.
To date, there is not yet a specific and effective antiviral agent against the SARS-CoV-2 available. Nevertheless, current research suggests that some treatments with appropriate viral restriction mechanisms may show promising results and approved methodological trials. The main protease of SARS CoV2 is a key protein that has an essential function in mediating viral replication and transcription. This enzyme is a prominent target for the development and the conception of antivirals against coronavirus infections.
The bibliographical screening for the lopinavir/ritonavir combination reports that clinical studies on SARS have been associated with reduced mortality and intubation rates, but their retrospective and observational nature precludes definitive findings1. Chloroquine or its hydrochloric derivative appears to be effective in preventing the replication of this virus; those molecules also play a particularly beneficial immunomodulatory function, which explains the implication of these molecules in the treatment of other autoimmune inflammatory diseases.
Other treatments were reported as therapeutic strategies against SARS Cov2 infections, notably Azythromycine2, oseltamivir, and lopinavir/ritonavir treatments were also adopted3-5 nitazoxanide, an antiviral against a broad range of viruses6 and Favipiravir7. In parallel and for a specific purpose, other molecules were used in this research, including molecules hypothesized to be involved in SARS CoV2 infections, the peptidomimetic AG7088, and drugs from the class of protease-targeting antivirals.
The objective of this research was to investigate the applicability of the SARS CoV2 main protease (ID: 6LU7) as a target protein for treatments by examining the binding and their affinities. Besides, this may provide insights into the current knowledge of the therapeutic efficacy of different pharmacological approaches.
The study selected the 3-chymotrypsin-like protease (3CL-protease) as a targeted enzyme, necessary for virus survival, which it’s role: the cleavage of polyproteins into replication-linked proteins. The 3CL-protease structure PDB ID: 6LU7 was obtained from the Protein Data Bank.
Referring to the literature data, we tested and analyzed the affinity of the recently reported pharmacological proposals. All the 3-D structures of these drugs were obtainable from the PubChem database. For the simulation of the binding affinity, and the structural visualization Molecular Operating Environment (Chemical Computing Group) had been employed.
The classification was based on the S score, permitting the assessment of the treatment and the active site of the protease affinities; meanwhile the Root Mean Square Deviation reference is used for the conformational analysis as well as the hydrophobic and polar interactions.
Table 1 presents the results of the molecules related to 3CL-protease. A part of these molecules is proposed and used. These scores were obtained from our modeling model and represent the relative binding affinity.
Table (1):
The docking score of molecules and the active site of Protease.
Molecules |
Therapeutic classification |
Score |
Root mean square deviation reference |
|---|---|---|---|
LOPINAVIR |
Antiretroviral protease inhibitor |
-9.3204 |
2.2137 |
AZITHROMYCINE |
Antibiotic |
-8.6789 |
3.2623 |
OSELTAMIVIR |
Antiviral neuraminidase inhibitor |
-7.2436 |
1.3627 |
HYDROXYCHLOROQUINE |
Anti-Infective Agents |
-7.2275 |
1.7114 |
TIPRANAVIR |
Antiviral nonpeptidic protease inhibitor |
-7.0007 |
1.6227 |
CHLOROQUINE |
Anti-Infective Agents |
-7.0001 |
1.0006 |
NITAZOXANIDE |
Anti-infective drug |
-6.5650 |
1.1217 |
AG7088 |
Peptidomimetic |
-6.1241 |
4.7793 |
REMDESIVIR |
Antiviral |
-6.0829 |
5.2271 |
ZANAMIVIR |
Antiviral Inhibit neuraminidase |
-5.8634 |
0.8937 |
NELFINAVIR |
Antiviral HIV-1 protease inhibitor |
-5.85685 |
2.6665318 |
SAQUINAVIR |
HIV protease inhibitor |
-5.6031 |
1.87757206 |
FAVIPIRAVIR |
Antiviral |
-4.6889 |
0.8465 |
Lopinavir had the highest binding affinity score, followed respectively by Azithromycin, Oseltamivir, Hydroxychloroquine, Tipranavir, Chloroquine, Nitazoxanide, AG70088, Remsidivir Zanamivir, Nelfinavir, Saquinavir and lastly Favipiravir.
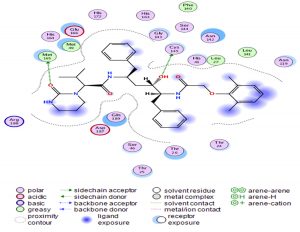
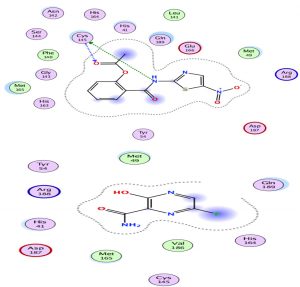
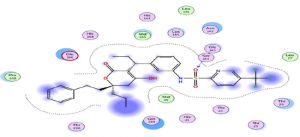
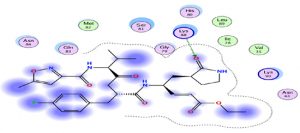
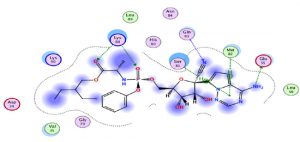
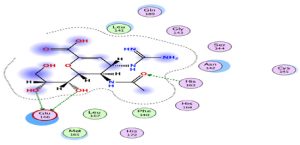
Through our methodological investigation on the analysis of the interactions between different molecules and the active site of the protease, we were able to reveal the sites of interaction between the different compounds and the residues involved in this interaction (Fig. 1). Through our structural interpretation and visualization of the different interactions, we have found that the amino acid Glycine 189 is the common element of the interaction of those treatments, which may be considered further to elucidate this result.
The characteristic of this interaction is the high polarity between Lopinavir and the active site of the Protease.
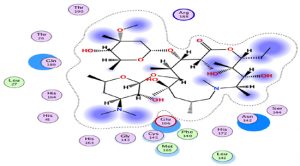
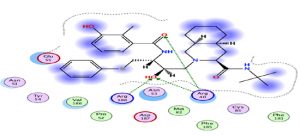
An interesting exploration, Azithromycin well-known for its anti-bacterial activity, showed an interesting result with a relatively high score compared to the rest of the treatments, hence it is interesting to investigate its clinical results. This interaction is characterized by a higher number of amino acids (Fig. 2).
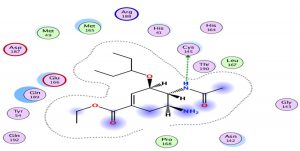
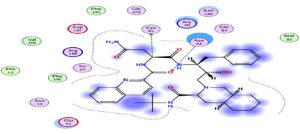
Oseltamivir as the first neuraminidase inhibitor available orally, classified in the third position with scores very close-to hydroxychloroquine, thus the interaction with the Protease is essentially with its N radical (Fig. 3).
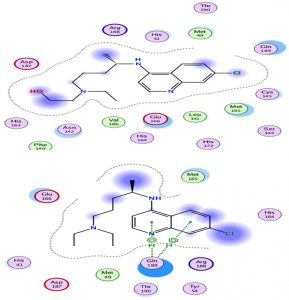
Chloroquine and its derivative have the same classification with an approximately identical score with hydrogen interactions. However, hydroxychloroquine seemed to be more effective than chloroquine (Fig. 4).
The lowest scores were observed for Sequinavir and Favipiravir respectively. It was also observed that Favipiravir had the lowest number of amino acids generated in this interaction (Fig. 5).
In the following table we provide the 3-dimensional interaction of the molecules that are not included in the therapeutic strategies proposed, although they might be used as therapeutic alternatives (Table 2).
Table (2):
3D illustration of interaction of molecules including antivirals drugs and peptidomimetic agent.
Molecules |
3D Illustration |
|---|---|
TIPRANAVIR |
|
AG7088 |
|
REMDESIVIR |
|
ZANAMIVIR |
|
NELFINAVIR |
|
SAQUINAVIR |
Remaining objective, our results demonstrate solely the scores that we were able to demonstrate based on our modeling methodology. That means that our results do not influence the favoritism of one treatment over the others. The treatments we have investigated in the manuscript are typically the subject of several clinical trials. Although our study is like the first trial, comparing the different treatments.
Hydroxychloroquine was reported to be of better affinity compared to Chloroquine8. These treatments are being proposed as a treatment for Covid19 and the clinical trials that may determine the effectiveness of these treatments are currently in progress9,10.
It has been suggested that the therapeutic effect of lopinavir on SARS-CoV2 may be primarily due to their inhibitory effect on endopeptidase11. Oral treatment with oseltamivir has been used as an empirical drug for several cases of SARS-CoV2 in Chinese hospitals12. Another analysis comparing only antiviral agents had mentioned their efficacy on the modeling level, this Screening has studied 61 molecules, Lopinavir binds to all SARS-CoV2 protein structures with a docking score less than -6.5. Also, it is interesting to note that HIV protease inhibitors have shown strong results in silico docking13.
A briefing in China reported that chloroquine has been used successfully to treat more than 100 SARS-CoV2 infected cases resulting in improved radiological findings and viral clearance, and reduced disease progression. The score we obtained throughout our docking can be explained by the indirect action on the virus, although the proposed mechanism for chloroquine and hydroxychloquine Increases endosomal and liposomal pH required for virus/cell fusion9.
All of our results provide an overview of the prescription and repositioning of the different treatments cited in the literature. It is important to notice that the benefit-risk ratio and the therapeutic tolerance range should be taken into consideration.
The azithromycin seems to have an inhibitive effect on viral replication, given its broad spectrum of activity. The documents mentioning azithromycin report that it only may be used in association with other treatments, whereby, based on our analysis, the second classified azithromycin would be considered as a treatment with adaptations according to its pharmacokinetic parameters. Oseltamivir is being used in several clinical trials as a comparison group; the reasons for this are not unraveled except its effectiveness against influenza. A recently published In silico research demonstrated that Oseltamivir may have an inhibitory effect on the SARS Cov2 main protease12.
There are limited trials investigating the antiviral activity against SARS Cov2 of the agents listed in Table 2. Concerning tipranaivir nonpeptidic protease inhibitor used in combination with ritonavir and demonstrated beneficial effect against HIV14. It was reported that nitazoxanide, through preventing membrane fusion and acting as an antiprotozoal agent, may have an inhibitory activity against COVID-1915. Zanamivir is an approved medication for the treatment of influenza A and B viruses16. Indinavir and Saquinavir have been shown to treat and prevent HIV. Remdesivir inhibits viral replication by initiating effective metabolic conversion in cells to activate nucleoside triphosphate which, in turn, inactivates viral RNA polymerases17. This agent has shown promising antiviral activity In-Vitro and is accessed for activity against SARS-CoV-218.
In the present context of the rapid spreading of the SARS CoV-2 pandemic and its huge impact both in terms of global health and socioeconomically, it is difficult to assess what would be the efficient alternative treatments in the shortest possible time. Although scientific rigor is inherent and adherence to guidelines is ideal, it seems difficult to achieve perfection and apply all methodological recommendations in every situation to evaluate and identify successful therapies.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declares that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. [published online February 27, 2020]. J Med Virol. 2020.
- P Gautret, J Lagier, P Parola, et al, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, International Journal of Antimicrobial Agents. 2020;105949,
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. doi :10.1016/ S0140-6736(20)30211-7
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;bv395:497–506. doi:10.1016/S0140-6736(20)30183-5
- Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020. [Epub ahead of print]. doi:10.1001/jama.2020. 1585.
- Wang, M, Cao R, Zhang L et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res., 2020; 30: 269–271.
- Q Cai, M Yang, D Liu, et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study, Engineering (2020),https://doi.org/10.1016/j.eng.2020.03.007
- Gonzalez-Paz LA, Lossada CA, Moncayo LS, et al. Theoretical Molecular Docking Study of the Structural Disruption of the Viral 3CL-Protease of COVID-19 Induced by Binding of Capsaicin, Piperine and Curcumin Part 1: A Comparative Study with Chloroquine and Hydrochloroquine Two Antimalaric Drugs. Research Square. 2020. DOI: 10.21203/rs.3.rs-21206/v1.
- Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi:10.5582/bst.2020.01047
- Andrea Cortegiani, Giulia Ingoglia, Mariachiara Ippolito, Antonino Giarratano, Sharon Einav, A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19, Journal of Critical Care, 2020.
- Shen Lin, Runnan Shen, Jingdong He, Xinhao Li, Xushun Guo. Molecular Modeling Evaluation of the Binding Effect of Ritonavir, Lopinavir and Darunavir to Severe Acute Respiratory Syndrome Coronavirus2 Proteases bioRxiv, 2020;929695
- Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J Biomol Struct Dynam. 2020; 1-7.
- Humi Shah, Palmi Modi, Sneha R Sagar, In silico studies on therapeutic agents for COVID-19: Drug repurposing approach, Life Sciences. 2020;252:117652.
- Doyon L, Tremblay S, Bourgon L, Wardrop E, Cordingley MG. Selection and characterization of HIV-1 showing reduced susceptibility to the nonpeptidic protease inhibitor tipranavir. Antiviral Research. 2005;68(1):27-35
- Yan Y, Shin WI, Pang YX, et al. The first 75 Days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment. International Journal of Environmental Research and Public Health. 2020;17(7):2323.
- LS Eiland, EH Eiland Zanamivir for the prevention of influenza in adults and children age 5 years and older. Therapeut Clin Risk Manag, 2007; 3(3): 461-465.
- World Health Organization. WHO R&D Blueprint—Ad-hoc Expert Consultation on Clinical Trials for Ebola Therapeutics. Available online: https://www.who.int/ebola/drc-2018/treatments-approved-forcompassionate-use-update/en/
- Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.