ISSN: 0973-7510
E-ISSN: 2581-690X
Stenotrophomonas maltophilia is a low-virulence opportunistic pathogen that causes human infections, especially in profound ill patients. Even if the bacterial genomes seem understood, the activities of many proteins are unknown. The purpose of our current research is to unravel the functional characteristics i.e. functional domain search and valuable regions of a hypothetical protein that would aid in the identification of potential drug targets in Stenotrophomonas maltophilia. The hypothetical protein of S.maltophilia was located and annotated using different in silico techniques. Our target protein was predicted to be Transcrip Reg superfamily YebC/PmpR based on motif and domain analysis by functional annotation tools. The regulator proteins of the YebC family are part of a vast collection of widely conserved hypothetical proteins with unclear functions. Examining and reviewing the function of YebC family protein, they repress Quorum sensing by directly binding to the promoter region of QS master regulator pqrS. It has also been reported that T3SS expression is regulated by YebC, to activate the virulence expression direct interaction with one of the T3SS promoters is needed.
Stenotrophomonas maltophilia, Transcrip Reg superfamily, hypothetical protein, virulence factor
Stenotrophomonas maltophilia is an organism with a complex taxonomic history. It is found worldwide and can live in a wide range of moist environments.1,2 These species can be isolated plant roots and soil, river water, tap water, vertebrates, and invertebrates.3 S. maltophilia is the third most pathogenic non-fermenting gram-negative bacillus (NFGNB) globally, causing severe human infections such as acute exacerbation of chronic obstructive pulmonary disease (COPD) and pneumonia. It also causes bloodstream infections (BSIs), urinary tract infections. Endocarditis and meningitis. It can cause persistent colonization and chronic infection in cystic fibrosis patients.4,5,6 Due to inbuilt resistance to a broad class of antibiotics, such as trimethoprim, fluoroquinolones, chloramphenicol, lactams, macrolides, aminoglycosides, cephalosporins, and polymyxin, new treatment techniques are necessary.7
The progress of genomics, particularly Next Generation Sequencing (NGS), allows us to gather a large amount of data in a short period8, 9 Statistical and algorithmic approaches play a vital part in the computational analysis of biological nucleotide sequences, which have become a very high subject of emerging research and a very interdisciplinary one10. More than 30% of proteins in many animals have unknown molecular functions, referred to as “hypothetical proteins (HP)”, which can be solved by in silico analysis. It aids in comprehending the organism’s DNA, which is necessary for the effective development of medication or vaccines. HP characterization using in silico methods aids in determining three-dimensional structures, which may disclose new pathways, domains and motifs, protein networks, and other information11,12,13 . Several bioinformatics databases and tools, including BLASTp, UNIPORT, ProtParam, CELLO, SOPMA, PSIPRED, SWISS PDB viewer, CASTp, Procheck, Autodock vina has been used to illustrate the activities of HP in a variety of infectious organism with great success.14,15
This study makes use of various in silico tools to screen and validate S. maltophilia functional proteome to interpret novel factors of virulence. The hypothetical protein was examined, annotated, and validated as transcrip reg Super family as a result of this study.
Selection of the hypothetical protein
The genome of S. maltophilia was sequenced and was submitted in NCBI (http:// www.ncbi.nlm.nih.gov/genome). The amino acid sequences in FASTA format of 1304 proteins which are classified as HP was acquired from a total set of 4707 proteins because their function and characteristics are unknown. Out of these 1304 HPs we have chosen one HP based on the results of BLASTP for sequence similarity and Protparam for stability check. HP with feature ID FIG|40324.1830. peg.444 (Supplementary Table 4) and 243 amino acid residues of Stenotrophomonas maltophilia strain was selected for further analysis.
Physiochemical property Analysis
Due to the unavailability of information on HP, such as AI (aliphatic index), GRAVY (Grand average of hydropathy), Isoelectric point, and extinction coefficients, Expasy’s ProtParam server16 was used to determine physiochemical parameters of a protein (pI).
Prediction of solubility and subcellular localization Following the prediction of the HP physiochemical qualities, the location of proteins was examined, which will aid in predicting drug and vaccination targets. CELLO17 calculated the average of hydrophobicity and determined the solubility of HP to estimate its subcellular location. The trans membrane region refers to any hydrophobic region of a protein.
Motif and domain analysis for function prediction NCBI CD Search (Conserved Domain Search Service) (https://www.ncbi.nlm.nih.gov/ Structure/cdd/wrpsb.cgi)18 InterProScan (https://www.ebi.ac.uk/interpro/ search/sequence/)19 and Pfam (http://pfam.xfam.org/)20 are all excellent resources for domain analysis. Proteins function is predicted using the Pfam database. CD Search detects the presence of conserved domains in the protein sequence. MOTIF server (https://www.genome.jp/tools/motif/) was used to examine the protein sequence motif.
Multiple sequence alignment
BLASTP8 search against the nonredundant database with parameters from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), an algorithm for predicting the similarity sequences of Jalview21 was utilized to create a phylogenetic tree and MSA.
Determining secondary structure
The secondary structure was predicted utilizing the SOPMA (https://npsa-prabi.ibcp.fr/ cgi-bin/npsa_automat.pl?page=/NPSA/npsa_ sopma.html) self-optimized prediction method with alignment9. PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/)10 was also used to validate the SOPMA results.
Homology model
Target protein’s 3D structure established using SWISS-MODEL21 server based on homology modeling. For each protein sequence, the server executes an automatic BLASTp search to find templates. The protein template chosen was 4f3q.1. A for the homology modeling based on the query result.
Energy minimization
YASARA14 force field minimizer was used to reduce the energy of a 3D structure from the SWISS-MODEL. It saves energy while producing a more correct and steady three-dimensional structure of a required protein.
Quality evaluation of the model
PROCHECK (https://saves.mbi.ucla.edu/)22, QMEAN23 (https://swissmodel.expasy.org/qmean/ help), Verify 3D24, and ERRAT25 were used to analyze the quality of the model structure. PyMOL26 overlaid and showed the model and template structure. ProSa web27 was also used to calculate the Z score.
Determination of Active site
Quantified mapping of the surface topography CASTp (http://sts.bioe.uic.edu/ castp/) was utilized to find the site of activation of the protein. CASTp aids in the discovery of a protein’s topographical features in an elaborated quantitative and comprehensive way. The three-dimensional structures can be measured appropriately and located14. Active pockets located on protein surfaces can also be measured. PyMOL software26 was used to visualize the CASTp output.
Comparative genome approach
BLASTp was used to see whether the target hypothetical protein is having any similarity with humans. BLASTp search (https://blast.ncbi. nlm.nih.gov/Blast.cgi)8 against the proteome of Homo sapiens was done. A minimal bit score of 100 and a threshold E-value of .005 were used for the filter hits.
Virulence protein prediction
To estimate the pathogenic ability and understand the complex virulence mechanism, it is essential to identify the virulent protein in the bacterial protein sequence. Here we used VirulentPredtool (http://203.92.44.117/cgi-bin/ virulent/chkres?22466)28 for the identification of virulent factor of the target protein. It is a Support Vector Mechanism (SVM) based method to predict the virulence protein with accuracy.
Molecular docking analysis
To verify the authenticity of the protein’s proposed function, molecular interaction tests of the proposed ligands were conducted. The interactions and binding affinity at the binding sites were regarded as the structurally annotated protein’s final functional annotations. Following that, the ligands binding interaction and affinity were compared to proteins from other genera in the database for structural and functional identity the binding pockets were identified using a prank web tool (https://prankweb.cz/). The grid box was determined by the binding site residues, with all binding site residues fitting within the grid box. The Discovery studio visualizer29 program was used to examine the molecular interactions in the complexes in great detail.
Ligand preparation
Using the ACS chem sketch tool, the inhibitors and ligand 3D structures were sketched. The geometrical alignment of the 3dimensional structure was checked. The 3-dimensional structure of ligands was then constructed by combining the respective 3D coordinates. The Argus tool was used to clean the geometry of the 3D structure that was thus created. For molecular docking investigations, the ligands improved 3D structure was used. The crystalline structure of protein obtained from the RCSB PDB was improved further by eliminating water residues, then adding Gasteiger charges, and finally joining nonpolar hydrogen utilizing Auto Dock V.4.030.
Molecular dynamics simulation studies
All unbiased MD simulations were performed with Gromacs -2016.3 simulation package. To diffuse the system, the TIP3P water model was employed, and NaCl counter ions were supplied to neutralize it. Using leap-frog integrator with a time step of 2 femtoseconds. The simulation was performed in an NPT ensemble. Nose Hoover Thermostat was used to maintain the average temperature of the system at 300K with the relaxation time of 1.0 PS and at 1 bar constant pressure with a coupling constant of 5.0ps respectively. The SwissParam webserver was used to create the ligand parameter files.
Physiochemical properties
Using the ProtParam tool, several physiochemical parameters of HP from S. maltophilia SMR013 strain were predicted to have 243 amino acids, a molecular weight of 25704.83, and a half-life of 30 hrs. It has a GRAVY of -0.17 and pI of 4.65 in terms of hydropathicity. The target proteins stability index was expected to be 26.00, indicating that it is stable (Supplementary Table 1).
Subcellular localization
Subcellular localization of an HP helps gain insight into its functions, as different cellular sites reflect distinct functions, which aids in drug development against a specific protein. The CELLO software predicted the target protein’s subcellular location as “cytoplasm”. The protein was predicted to be a soluble protein by the SOSUI server.
Phylogeny and protein family analysis
NCBI-CD Search, InterProScan, and Pfam were used for prediction. They suggested containing the domain of the Transcrip reg superfamily. NCBI-CDD server detected that Transcrip reg superfamily region at 7-243 amino acid (aa) with 2.93e-121 E-value. Pfam detected the transcrip reg superfamily domain at 5-235 amino acid with an E value of 2.0e-87. MOTIF found like transcrip reg superfamily at 7-235 position with 1.3e-86 E-value (Supplementary Table 2).
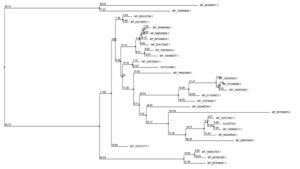
BLASTP analysis against the nonredundant database revealed sequence similarity of up to 92 percent. Supplementary Fig. 1 shows an MSA of four amino acid sequences from the result of BLASTp. Jalview software was used to create a phylogenetic tree utilizing numerous protein sequences. The protein employed to create the phylogenetic tree were primarily from S. maltophilia (Fig. 1).
Fig. 1. Phylogenetic tree depicting the target proteins’ evolutionary relationship to other transcription regulatory proteins. Jalview program constructed the tree using the neighbor-joining approach relative to the BLOSUM62 scoring matrix. The mismatch of the percentage of 2 different nodes (branch length) is represented by this value.
Model analysis and quality evaluation
The protein’s secondary structure was predicted using SOPMA and PSIPRED. The alpha helix was determined to be the most prevalent (47.74) following the random coil (27.57), extended strand (15.23), and beta-turn (9.47). PSIPRED came up with similar results (alpha-helix: 45.67%, random coil: 34.58%, and extended strand: 19.75%). PSIPRED predicts the proteins’ secondary structure (Supplementary Fig. 2).
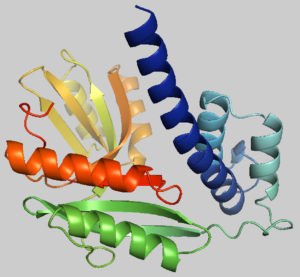
Fig. 2. Predicted 3-dimensional structure of the target protein through SWISS-MODEL server after YASARA energy minimization.
The protein tertiary structure was determined using the template 4f3q.1.A from the SWISS-MODEL server, which shares 54.77 % sequence similarity with the desired protein. Fig. 2 shows the model that was produced using SWISS-MODEL.
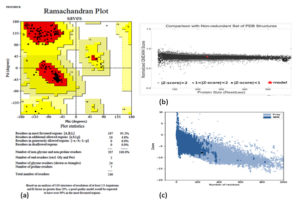
PROCHECK, Varify 3D, ERRAT, and QMEAN were utilized to examine the quality level of the modeled three-dimensional structure. According to PROCHECK results, 95.2 % of amino acid residues in the “Ramachandran plot” fall into the most liked zone (Fig. 3). The model passed the Verify 3D server with an average 3D-1D score >0.2 for 8625 percent residues. The model was placed into the dark grey zone by the QMEAN tool. The value of QMEAN4 is 0.80, which is deemed suitable. The structure of the protein was predicted as good quality by ERRAT, 97.3333 of quality factor.
Fig. 3. Evaluation of models quality: (a) Ramachandran plot of model structure validated by PROCHECK program, (b) Graphical representation of QMEAN result of the model structure (indicates good agreement between model structure and experimental structures similar size and (c) Z- score of the protein using proSA server.
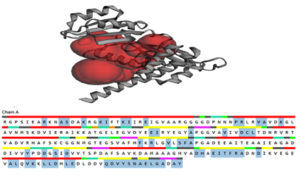
Fig. 4. The active site was determined by the CASTp server, which was then represented in PyMOL as two big pockets (up). In the diagram below, the active amino acid residues have been highlighted.
The model’s Z score measures the overall quality of the model and is used to see if the input structure falls inside that range of scores found in native proteins that are similar. The model obtained with ProSA had a Z score of -7.86.
Active site determination
CASTp server was used to analyze the active site of the model structure and determine the active site amino acid residues. PyMOL was then used to visualize the outcome (Fig. 4). The first stage in designing a medication or inhibitor is to identify and characterize active site residues. As per CASTp prediction, model proteins active residues (of the 2 most significant active pockets with SA area of 3200.932 and 1559.820 respectively) are Arg45, Ala13, Ser14, Lys24, Ils25, Glu28, Pro44, Val94, Asp97, Cys98, Leu99, Phu131, Tyr163, His194, Thr198, Phe199, and Gln229.
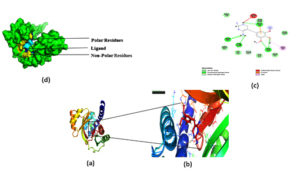
Fig. 5. The docking site and interaction were highlighted in (a, b).which was then represented in PyMOL,(c ) 2D diagram of the ligand and interacting residues in the protein. (d) Surface plot of the protein, polar residues was highlighted in yellow non-polar residue highlighted in Blue and ligand was highlighted in cyan.
Fig. 7. Showing the analysis of RMSD, Rg of total axis, Rg of individual axis and SASA graphs of hypothetical protein and ligand.
Comparative Genomic Approach
To investigate if the target protein had human homologues, BLASTp search was run against the human proteome. 4 similar sequences were found in which Sequence id EAW94302.1 has 30% identity and query cover of 44%.
Virulence factor
Infection is established by pathogens with the help of Virulence factors. The virulence factor was studied using VirulentPred. The creation of novel medications could be aided by understanding these dangerous proteins. The protein is virulent, and the predicted score is 1.0595.
Molecular Docking
The P2Rank approach is based on the categorization of points equally distributed over the Solvent Accessible Surface of proteins (referred to as SAS points). Local spherical 3D neighborhoods centered on these locations are represented by these points. At the same time, they can be seen as possible contact atom sites for prospective ligands. SAS points are first characterized by a vector of physio-chemical, geometric, and statistical characteristics derived from their immediate geometric surroundings. A machine learning-based approach assigns a projected ligand ability score to each SAS point in turn. Finally, high estimated ligand ability spots are grouped to create anticipated ligand binding sites. The high-rank score binding pockets were taken for Docking.
Docking was performed on the binding pocket with the highest rank score. The predicted hypothetical protein binding site’s coordinates were X-15.82, Y-16.57, and Z-59.37, and Trimethoprim interacted with it through hydrogen bonds at ASP95, LYS50, TYR230, GLU26, alkyl bonds at VAL46, unfavorable donor interaction at ARG43, and van der Waals interaction at ASP228. The binding affinity was found to be -5.8 kcal/mol in Fig. 5: a,b,c.
Molecular Dynamics Studies
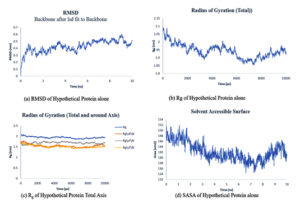
The simulation period and protein variation, as well as their complexes, are examined using the root mean square deviation (RMSD), the radius of gyration (Rg), solvent accessible surface area (SASA), and the number of hydrogen bonds maintained during the simulation study. For a set amount of time, the simulation was conducted. Using the natural protein alone and in combination with a 10 ns ligand.
The RMSD plots of hypothetical protein alone and ligand protein complex Fig. 6: (a) and Fig. 7: (b) attained equilibrium at 8-10 ns and remained unstable with a variation of 0.3-0.5nm and RMSD range.
The gyration radius takes into account the various masses when calculating the root mean square distance using the central axis of rotation. Throughout the simulation, the Rg plots Fig. 9:(a) and Fig. 10: (b,c) considered shape and folding at each time step of the entire journey. The protein and its ligand complex have a similar Rg value trend, with a variance of 1.91-2.1nm for the hypothetical protein alone. For the 2.1-2.4nm complex.
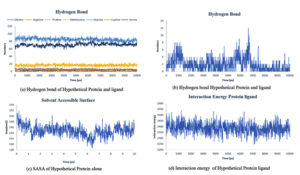
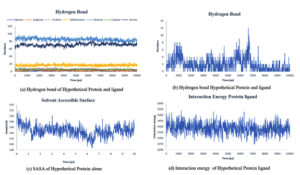
Fig. 8. Showing the analysis of (a) Hydrogen bond of protein ligand interaction respect to individual amino acids, (b) Total hydrogen bond of hypothetical protein ligand complex (c) SASA of hypothetical protein (d) Interaction energy between the protein and ligand.
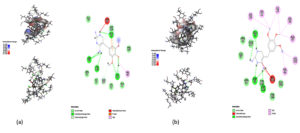
Fig. 10. Showing the analysis of Hydrogen bond of protein ligand interaction respect to individual amino acids, Total hydrogen bond of hypothetical protein ligand complex SASA of hypothetical protein Interaction energy between the protein and ligand. (a) Zero Nanosecond (b) ten Nanosecond.
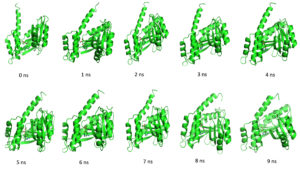
Throughout the simulation, H-bonds that arise during the Molecular Docking study are explored. All intermolecular H-bonds between ligands and proteins were only taken into consideration and displayed during the study. Fig. 8: (a,b). The plot illustrates that the number of H-bonds created during simulation runs matches the Molecular Docking study, with just a few bonds broken and repaired at the same time. SASA determines the region surrounding the protein and surface system-formed hydrophobic core. The values of SASA were discovered to be consistent. During the modeling procedure Fig. 8: (c). The interaction energy between the protein and the ligand was stable in the entire 10 ns. Fig. 8: (d). and every one nanosecond protein structure was shown in Fig. 9. Trimethoprim interacted with it through hydrogen bonds at ASP95, LYS50, TYR230, GLU26, alkyl bonds at VAL46, unfavorable donor interaction at ARG43, and van der Waals interaction at ASP228 in 0 nanoseconds and 10 nanoseconds the trimethoprim interact with hydrogen bond GLU26, ASP95, ASP228, and non-favourable bond between NA ions in atom position 7655 and van der Waals force through ARG43, LEU97, ARG81, THR 196 and GLU194 (Fig. 10). The interaction energy between the protein-ligand complex was stable showed in Fig. 8:(d).
An HP is a protein for which there is a lack of experimental evidence that it is expressed in vivo but whose presence has been predicted. Extensive genome sequencing has resulted in several predicted open reading frames to which functions cannot be readily assigned (Galperin, 2001). Characterization of hypothetical proteins can help to understand bacterial metabolic pathways, disease progression, drug development, and disease control strategies.
Various bioinformatics methods were employed to characterize the function and structure of the HP- feature ID FIG|40324.1830. peg.444 of the S. maltophilia SRM 01 strain in this investigation. The protein was calculated to comprise 243 amino acids with a molecular weight of 25704.83, 4.65 of theoretical pI, GRAVY of -0.172 based on physiochemical parameters (Supplementary Table 1). This protein was predicted to be extracellular by the CELLO server. Random coil, beta-turn, alpha helix, and extended strand make up the proteins’ secondary structure, with the most prominent the random coil. CD Search, Pfam, and InterProScan suggested that our HP belongs to a Transcrip reg superfamily named YebC/PmpR based on motif and domain analysis. PmpR is a member of the YebC family and subsequently binds to the pqsR promoter thereby negatively regulating PqsR expression and AQ production (Liang et al., 2008). This is a transcriptional regulator family. It induces the transcription of mitochondrially encoded COX1 in animals. It binds to the promoter region of the quorum-sensing response regulator in bacteria, which inhibits it. When a minimum level stimulatory quantity of an auto inducer is detected, gene expression changes. The prediction was validated by BLASTp result against the nonredundant database, which revealed up to 92 % sequence identity with other YebC/pmpR family DNA binding transcriptional regulators (Table 2). It includes Xanthomonas sp. (WP 166923906.1), Pseudomonas aeruginosa (WP 093295939.1), and, Pseudoxanthomonas Mexicana (WP 187574431.1) which has a similar protein. In Pseudomonas aeruginosa it binds to the promoter region of the Pseudomonas quinolone signal system’s quorum sensing response regulator pqrS, which has a detrimental effect on it. In most cases, Stenotrophomonas maltophilia seems to harm the quorum sensing system of its competitors. This is since S. maltophilia isolates have a diverse set of quorums quenching mechanisms, including the synthesis of fatty acids with quenching properties and the degradation of N-acyl homoserine lactone and palmitic acid methyl ester.30 The proteins 3D structure obtained using the SWISS-MODEL server fulfilled all the model qualities checked using PROCHECK, ERRAT, Verify 3D, and QMEAN.
After YASARA (energy minimization process), the three-dimensional structure became more stable. CASTp server was analyzed the active site of the model structure and determined the active site amino acid residues. The first stage in designing a medication or inhibitor is to identify and characterize active site residues. As per CASTp prediction 2 most significant active pockets with SA area of 3200.932 and 1559.820, respectively, were viewed with the help of PyMOL. A comparative genomic approach was checked to see whether the target protein would have had human homologues. Sequence id EAW94302.1 has 30% identity and query cover of 44%. The trimethoprim with good binding affinity as indicated by the binding site of the hypothetical protein of Stenotrophomonas maltophilia. VirulentPred was used to check the pathogenicity, which confirmed that the organism is virulent, and the predicted score is 1.0595.
Our work has been entitled to generate the first Hypothetical protein annotation in Stenotrophomonas maltophilia which plays a multitude of roles in different organisms. It was shown that this Transcrip reg superfamily named YebC/PmpR is involved in resolving the Holliday junction during the occurrence of homologous recombination event for chromosome segregation. It directly regulates RuvABC11. It was again demonstrated that YebC regulates variable surface antigen VlsE expression and is required for host immune evasion in Borrelia burgdorferi31. Later in 2021 Patil and his group also revealed that this transcription factor superfamily has a significant role in the growth of S. maltophilia at human body temperature as the transcriptomic analysis revealed around 2.5-fold change (downregulation) in gene expression of this transcription factor at 37°C when compared to 28°C.32 Wei and his colleagues displayed the role of YebC in regulating quorum sensing and virulence by triggering T3SS gene expression in the pathogen Edwardsiella piscicida.33 It was also exhibited that in the metal-tolerant bacterium Alishewanella sp., the YebC family protein is involved in As3+, Cr6+, and Cd2+ resistance.34 Thus in Stenotrophomonas maltophilia we assume that it could be involved in the regulation of quorum sensing and virulence as we have discussed above.
The uncharacterized hypothetical proteins block the hunt for new and more potent medication targets for pathogenic living things, as well as progress toward developing research on these forms of life and improving our understanding of their pathogenicity and virulence. The findings of this study aid in the understanding of the activities of HP of Stenotrophomonas maltophilia. The physiochemical properties, subcellular localization, function prediction, secondary structure prediction, MSA, 3D structure, virulence checking, active site determination, and molecular docking done in this study will help develop drugs and future studies about the target protein.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Brooke JS. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25(1):2-41.
Crossref - Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11(1):57-80.
Crossref - Berg G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84(1):11-18.
Crossref - Lipuma JJ, Currie BJ, Peacock SJ, Vandamme PAR. Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia and Acidovorax. In: Manual of Clinical Microbiology. ASM Press; 2015:791-812.
Crossref - Trifonova A, Strateva T. Stenotrophomonas maltophilia–a low-grade pathogen with numerous virulence factors. Infect Dis (Auckl). 2019;51(3):168-178.
Crossref - Waters V, Atenafu EG, Lu A, Yau Y, Tullis E, Ratjen F. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J Cyst Fibros. 2013;12(5):482-486.
Crossref - Peters DL, Lynch KH, Stothard P, Dennis JJ. The isolation and characterization of two Stenotrophomonas maltophilia bacteriophages capable of cross-taxonomic order infectivity. BMC Genomics. 2015;16(1).
Crossref - Xu J -H. Basic Local Alignment Search Tool. In: Catalysis from A to Z.; 2020.
Crossref - Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network protein sequence analysis. Trends Biochem Sci. 2000;25(3):147-150.
Crossref - Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402-W407.
Crossref - Idrees S, Nadeem S, Kanwal S, et al. In silico sequence analysis, homology modeling and function annotation of Ocimum basilicum hypothetical protein G1CT28_OCIBA. Int J Bioautomation. 2012;16(2):111-118. Accessed August 9, 2021. http://biomed.bas.bg/bioautomation/2012/vol_16.2/files/16.2_02.pdf
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96(8):4285-4288.
Crossref - Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189-1191.
Crossref - Krieger E, Joo K, Lee J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct Funct Bioinforma. 2009;77(SUPPL. 9):114-122.
Crossref - Mishra R, Bijarnia-Mahay S, Kumar P, et al. Early Infantile Thiamine Transporter-2 Deficiency with Epileptic Spasms—A Phenotypic Spectrum with a Novel Mutation. J Pediatr Epilepsy. Published online June 24, 2021.
Crossref - Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531-552.
Crossref - Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins Struct Funct Genet. 2006;64(3):643-651.
Crossref - Lu S, Wang J, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020. – PubMed – NCBI. Nucleic Acids Res. 2020;48(D1):D265-D268. Accessed August 13, 2021. https://academic.oup.com/nar/article-abstract/48/D1/D265/5645006
- Quevillon E, Silventoinen V, Pillai S, et al. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33(SUPPL. 2).
Crossref - Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2000;28(1):263-266.
Crossref - Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296-W303.
Crossref - MacArthur MW, Laskowski RA, Thornton JM. Knowledge-based validation of protein structure coordinates derived by X-ray crystallography and NMR spectroscopy. Curr Opin Struct Biol. 1994;4(5):731-737.
Crossref - Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27(3):343-350.
Crossref - Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396-404.
Crossref - Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511-1519.
Crossref - Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44(5):433-437.
Crossref - Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(SUPPL.2).
Crossref - Garg A, Gupta D. VirulentPred: A SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinformatics. 2008;9.
Crossref - Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera – A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-1612.
Crossref - Shin WH, Heo L, Lee J, Ko J, Seok C, Lee J. LigDockCSA: Protein-ligand docking using conformational space annealing. J Comput Chem. 2011;32(15):3226-3232.
Crossref - Zhang Y, Chen T, Raghunandanan S, et al. YebC regulates variable surface antigen VlsE expression and is required for host immune evasion in Borrelia burgdorferi. PLoS Pathog. 2020;16(10).
Crossref - Patil PP, Kumar S, Kaur A, Midha S, Bansal K, Patil PB. Global transcriptome analysis of stenotrophomonas maltophilia in response to growth at human body temperature. Microb Genomics. 2021;7(7).
Crossref - Wei L, Wu Y, Qiao H, et al. YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol Lett. 2018;365(14).
Crossref - Wu S, Xia X, Wang D, Zhou Z, Wang G. Gene function and expression regulation of RuvRCAB in bacterial Cr(VI), As(III), Sb(III), and Cd(II) resistance. Appl Microbiol Biotechnol. 2019;103(6):2701-2713.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.