ISSN: 0973-7510
E-ISSN: 2581-690X
Dodonaea viscosa Jacq is known as “chamana” in the popular flora of Peru. The traditional medicine uses its leaves as ingredient in fermented beverages from Zea mays and also in external uses for anti-inflammatory diseases. The aim was to study the role of dodonic acid against several protein targets of S. aureus. This study was focused on to analyse the role of dodonic acid against S. aureus target proteins such as on Sortase-A, DNA gyrase, dihydrofolate reductase (DHFR), clumping factor, dehydrosqualene synthase, and undecaprenyl di-phosphate synthase as a promising candidate molecule. The docking analysis of dodonic acid showed the best docking score energy on S. aureus undecaprenyl diphosphate synthase with -11.2 kcal/mol and demonstrated to be a very stable molecule at physiological conditions during the molecular dynamic for 50 ns. As conclusion, the extract demonstrated to be active against S. aureus and dodonic acid might be a promising molecule acting on the S. aureus undecaprenyl diphosphate synthase.
Dodoviscin, antibacterial, docking analysis, molecular dynamic, medicinal plants, in-silico
Peru is one of the megadiverse countries in the world with a wide variety of medicinal plants distributed in its three regions (coast, mountain and rain forest), many of them have been screened as antibacterial drugs in vitro, based on the ethnobotanical and ethnopharmacological information selected gram-negative and gram-positive bacteria were used to find any promising activity.1-3 However, in a study reported by Bussmann of 166 plants only 32% showed any antibacterial activity within this plants, some are used for diarrhea, respiratory disorders, inflammation, infections, as well as in complications during post-partum infections.4 On the other hand, in the mountains of Apurimac (a region from Peru) grow a medicinal plant known as “shamana”, which is scientifically known as Dodonaea viscosa, in which leaves are used for magical purposes and also healing for inflammatory disorders and body pain.5 Additionally, it is known that the species in question can be found in different parts of the world such as in the Asian continent where in countries such as India and China it is considered part of their traditional medicine, being used in decoctions and infusions and for the treatment of diarrheal diseases, ulcers and inflammatory-type disorders.6-8
The Sapindaceae family contains around 1000 species. Most of them are woody and flowering plants used to treat different diseases including heart disease. Dodonaea viscosa Jacq (D. viscosa) is distributed in particular regions of Africa, Mexico, Australia, United States, South America countries, and elsewhere.9 Regarding the biological activities of D. viscosa, different extracts have demonstrated antiproliferative effect against various types of tumor cell lines A2780 (ovarian), BT-549 (breast), DU 145 (prostate), NCI-H460 (lung), and HCC-2998 (colon),10 antioxidant, cytotoxic, antidiabetic and antimicrobial,11 antiinflammatory,12 cytotoxic against HT-29 colon cancer cells,13 anti-diarrheal 7 among other. These effects, even at an early stage, are probably attributed to its chemical components such as flavonoids, alkaloids, diterpenoids, saponins, tannins, steroids, proteins, carbohydrates, etc. Some compound isolated from leaves were named 3,3′,4′,5,7-pentahydroxy flavane, 4-methoxylstigmasterol, 5-hydroxy-7,3′4′-trimethoxy-6-acetoxy-3-prenylflavone, and dodonic acid.14 About dodonic acid, in an in vitro study showed effect on Staphylococcus aureus at 250 µg/mL.15
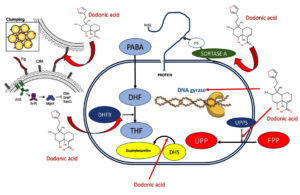
Currently, S. aureus is considered a multidrug-resistant microorganism, which has widespread worldwide. This bacteria can produce since serious skin infections to fatal pneumonia and sepsis. The incidence rate is found between 20 to 50 cases/100,000 per year, whilst in the United States the annual number of death was 20,000 by 2017. Lately, S. aureus is associated to atopic dermatitis and current antibiotics are being ineffective to combat this infection. In spite of the development of new drugs for S. aureus, the microbial infections are increased due to multidrug-resistant microorganisms overall in those acquired in hospitals.16 In this study, the aim was to determine the antibacterial activity in silico of dodonic acid against target proteins of S. aureus using a molecular modeling on dihydrofolate reductase (2W9S), Sortase-A (1T2P), DNA gyrase (3U2D), clumping factor A (1N67), undecaprenyl diphosphate synthase (4H8E) and dehydrosqualene synthase (2ZCO) (Figure 1).
Figure 1. Proposed action mechanism of dodonic acid against several targets of S. aureus. PABA: pa-ra-aminobenzoic acid; HF: dihydrofolate; THF=tetrahydrofolate; DHFR: dihydrofolate reductase; DHS: dehydrosqualene synthase; FPP: farnesyl pyrophosphate; UPPS: Undecaprenyl diphosphate synthase; UPP: undecaprenyl diphosphate product.
Molecular Docking of dodonic acid and S. aureus proteins
The main compound found in the ethanol extract of D. viscosa named dodonic acid was analyzed against S. aureus target proteins (PDB IDs: 4H8E, 1N67, 1T2P, 2W9S, 2ZCO and 3U2D). Molecular docking studies were performed using Autodock v 4.2.6. Binding cavity for the dodonic acid in S. aureus proteins were determined from the predefined co-crystallized X-ray structure from RCSB PDB. The residue positions were calculated within 3 Å space from the co-crystallized ligand. After the cavity selection in each case, the co-crystallized ligands were removed using Chimera tool (https://www.cgl.ucsf.edu/chimera/) and subsequently energy was minimized using steepest descent and conjugate gradient algorithm. Then finally, merging the nonpolar hydrogens, both receptor and target compounds were saved in pdbqt format. Grid boxes were created with specific dimensions in 0.3 Å spacing. Following the Lamarckian Genetic Algorithm (LGA), docking studies of the protein-ligand complex were performed to achieve the lowest free energy of binding (∆G). During molecular docking studies, three replicates were performed having the total number of solutions computed 50 in each case, with population size 500, number of evaluations 2500000, maximum number of generations 27000 and rest the default parameters were allowed. After docking, the RMSD clustering maps were obtained by re-clustering with a clustering tolerance 0.25 Å, 0.5 Å and 1 Å, respectively, in order to obtain the best cluster having lowest energy score with high number of populations. All docking parameter setting was done as described elsewhere. 17,18
Molecular Dynamics studies of dodonic acid and S. aureus proteins
The MD simulation was carried in order to analyze the stability of dodonic acid and S. aureus undecaprenyl diphosphate synthase (PDB ID: 4H8E) complex for 100 ns using the Desmond 2020.1 from Schrödinger, LLC. The OPLS-2005 force field,19,20 and explicit solvent model with the SPC water molecules were used in this system with a periodic boundary condition in an orthorhombic box and having dimension 10Å x 10Å x 10Å.21 Na+ ions were added to neutralize the charge. 0.15 M, NaCl solutions added to the system to simulate the physiological environment. Initially, the system was equilibrated using NVT ensemble for 100 ps to retrain over the protein-Aptamer complex. Followed by this a short run equilibration and minimization using NPT ensemble for 12 ps. The NPT ensemble was set up using the Nose-Hoover chain coupling scheme22 with temperature 300 K, the relaxation time of 1.0 ps and pressure 1 bar maintained in all the simulations. A time step of 2 fs was used. The Martyna-Tuckerman–Klein chain coupling scheme23 barostat method was used for pressure control with a relaxation time of 2 ps. The particle mesh Ewald method24 was used for calculating long-range electrostatic interactions, and the radius for the coulomb interactions were fixed at 9Å. RESPA integrator was used to calculate the non-bonded forces. The root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), B-factor fluctuation of C-α backbone of protein and number of H-bonds formed between acid and S. aureus undecaprenyl diphosphate synthase, were employed to monitor the stability of the MD simulations.
Molecular Docking of dodonic acid and S. aureus proteins
Target proteins from S. aureus interacting with dodonic acid were studied to establish the possible molecular mechanism involved in the antibacterial activity against S. aureus. Hence, some targets such as sortase-A (PDB ID: 1T2P), which is exclusively found in gram-positive bacteria and considered a type II membrane protein), it has a role in the cell wall envelope assembly.25 S. aureus DNA gyrase belongs to a subclass of type II topoisomerase is essential in DNA replication as well as in elongation and transcription.26 S. aureus clumping factor A (ClfA) (PDB ID: 1N67) is a class of adhesins involved in the initiation of infection and main virulence factor in S. aureus.27 The protein dihydrofolate reductase (2W9S) is involved in the production of tetrahydrofolate (THF) and its inhibition produces the destruction of the intracellular tetrahydrofolate pool 28. In addition, the dehydrosqualene synthase is an enzyme that has a protective role in the oxidative stress produced by the host immune by producing a pigment named staphyloxanthin.28 S. aureus undecaprenyl diphosphate synthase (UPPS) (PDB ID: 4H8E) leads to formation of peptidoglycan and only is not present in humans.29
Table:
Binding energy predicted inhibitory concentration profile and residue interactions between dodonic acid and S. aureus proteins.
Complex |
Binding energy (ΔG) [kcal/mol] |
Ki [nM] |
Polar contact interactions |
Non-bonded interactions |
|---|---|---|---|---|
1N67 clumping factor A |
-9.6 |
21 |
Val450, Phe449, phe445, Val490, Thr397, Tyr448, Arg395, Ser447, Pro251, Val288, Tyr369 |
His252, Ile488, Tyr399, Pro341 |
1T2P Sortase-A |
-8.6 |
31 |
Asn107, Thr93, Gly192, Ala92, ALA118, Trp194, Val193, Arg197, Glu105, Glu106, Asn107 |
Pro91, Ile182, Tyr187 |
2W9S dihydrofolate reductase |
-8.6 |
31 |
Thr96, Lys45, Arg44, Gln65, Thr63, Asn64 |
Leu62, Ala43, Ala100, Leu97, leu79 |
2ZCO dehydrosqualene synthase |
-8.0 |
40 |
Ile86, Avg85, Lys13, Tyr10, Lys46 |
Lys17, Ile14, Avg45, Asp49 |
3U2D DNA gyrase |
-9.3 |
24 |
Glu193, Gln197, Thr194, His46, Glu41, Arg42 |
His45, Trp49, Arg198 |
4H8E undecaprenyl diphosphate synthase |
-11.2 |
11 |
Arg201, Ile31, Asp33, Asn35, Gly53, Ile57, Met54, Leu95, Asn81, Ser78, Arg84, Phe77 |
Tyr75, His50, Met32, Ala76, Ile92 |
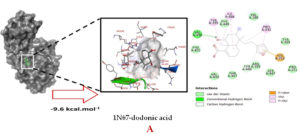
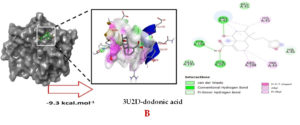
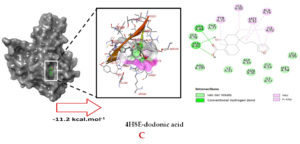
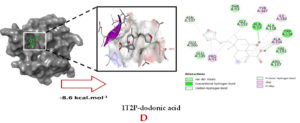
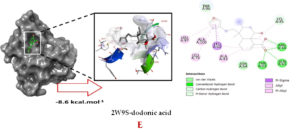
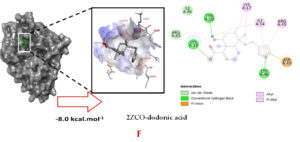
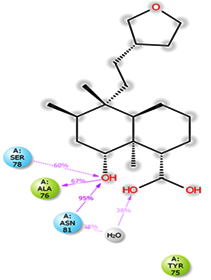
Molecular docking studies were performed to decipher the binding aspects of dodonic acid-with the S. aureus proteins (PDB IDs: 4H8E, 1N67, 1T2P, 2W9S, 2ZCO and 3U2D). The images of docked complexes, molecular surfaces, 3D and 2D interactive plots for dodonic acid with the proteins of S. aureus are shown in Figure 2. Molecular docking studies revealed that the dodonic acid bound significantly with S. aureus protein 4H8E with lowest binding energy -11.2 kcal/mol and with an inhibitory concentration (Ki) 11 nM. On the other hand, dodonic acid formed pi-cation interaction with His252, conventional hydrogen bond with Val450 residues of clumping factor A (Figure 2A). Other non-bonded interactions are depicted in Table. S. aureus Sortase-A (1T2P) and dydrofolate reductase bound (2W95) to dodonic acid had less affinity as compared with S. aureus protein 4H8E (un-decaprenyl diphosphate synthase) with a lowest binding energy -8.6 kcal/mol and the predicted Ki was found to be 31 nM. In this interaction, dodonic acid formed a conventional hydrogen bond with Ala92 and Trp194 residues (Figure 2C). On the other hand, S. aureus protein 2W9S bound with dodonic acid with same binding energy -8.6 kcal/mol and the predicted Ki was found to be 31 nM. Therefore, both 1T2P and 2W9S has same affinity for dodonic acid. The conventional hydrogen bond formed between dodonic acid and the amino acid residues of 2W9S, Arg44, Thr63 and Gln65 (Figure 2D and 2E). With 2ZCO, dodonic acid displayed poor binding with binding energy (ΔG) -8.0 kcal/mol and Ki 40 nM. Tyr10, Lys13 and Lys46 involved in the conventional hydrogen with dodonic acid
(Figure 2F). S. aureus protein 3U2D displayed significant binding with dodonic acid with -9.3 kcal/mol and having predicted Ki 24 nM. Conventional hydrogen bonds were observed between the ligand and Arg42, Thr194 and Gln197 (Figure 2B). However, S. aureus undecaprenyl diphosphate synthase (PBD: 4H8E) displayed high affinity for ligand with lowest binding energy (ΔG) -11.2 kcal/mol with lowest concentration of predicted inhibition (Ki) 11 nM. Conventional hydrogen bonds are formed between dodonic acid and the binding cavity residues of undecaprenyl diphosphate synthase Arg84, Ser78 and Asn81 (Figure 2C). Other non-bonded interactions are listed in Table. In a study of Ragi et al 28, Schiff base compounds has more affinity by 2W9S from S. aureus in sites 2 and 3 of the target (− 10.5 and − 9.5 kcal/mol) and interacted mainly with amino acid residues LEU20, ILE50, and PHE92, which were the main sites actives of this compound. However, our findings differ of those results because our compound had lees affinity by 2W9S (− 8.6 kcal/mol).
Figure 2. Molecular interactions studies of dodonic acid with the S. aureus proteins (A) 1N67, (B) 3U2D, (C) 4H8E, (D) 1T2P, (E) 2W95 and (F) 2ZCO surface view (Left panel) and 2D (Right panel) interactions.
On the other hand, extracts of different polarities were tested as antibacterial against several strain of S. aureus, in a study of Khurram et al., which used the crude extract and four different fractions, the response against S. aureus was negative with 12.0 ± 0.3 mm of inhibition zone 30. Bimani et al., only found antibacterial activity against S. aureus in methanol extract at 2 mg/mL with 13 ± 0.15 mm of inhibition zone. The n-hexane, butanol, ethyl acetate, chloroform, and water extracts did not have any effect with inhibition zone less than 5 mm. However, the isolated compound named 3, 5, 7, 3′,4′-pentahydroxyflavone (quercetin) had an activity of 15.00 ± 0.23 mm, being more than the methanol extract and the second abundant metabolite named hautriwaic acid which is a diterpenoid seemed to dodonic acid.31 On the other hand, Priya et al., demonstrated that the diethyl ether extract at 15 mg/disc had the better effect compared to the other extract such as ethyl acetate, acetone, and ethanol. These studies stated that the antibacterial effect could be due to the polar components instead of non-polar components.32 On the contrary, other repots referred to water extract with the best effect against S. aureus with 10 mm of inhibition zone at 0.25 mg/mL.33 Additionally, the ethyl acetate extract of D. viscosa had a better effect at 4 mg/mL with 18 mm of inhibition zone.34 Other, the ethyl acetate fraction at 100 µL/disc showed 16.1 ± 0.14 mm.35 According to the results of Daniel et al., the nanoparticles based on reduction of D. viscosa extract leaves might increase the antibacterial activity, the study evaluated three types of nanoparticles such as copper, zerovalent iron (ZVI), and silver nanoparticles, the results showed that ZVI had an effect of 14 mm at 10 µg/disc and 27 mm for 100 µg/disc.36
Molecular Dynamics studies of dodonoic acid and S. aureus proteins
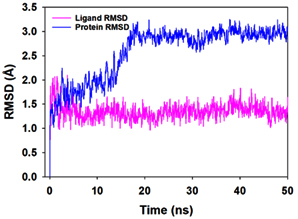
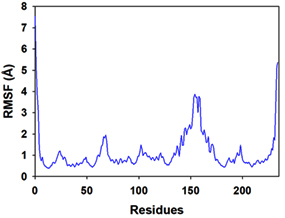
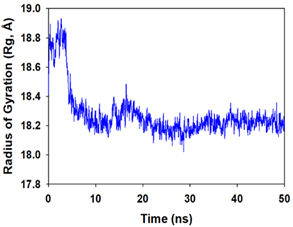
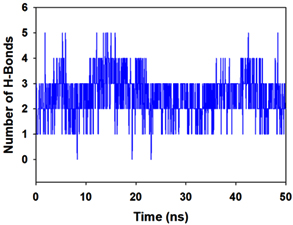
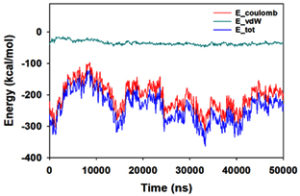
Molecular dynamics and simulation (MD) of the dodonic acid bound S. aureus undecaprenyl diphosphate synthase (PDB ID: 4H8E) was studied in detail to understand the nature of possible binding motifs and structurally stable conformations. The root mean square devia-tion (RMSD) of C a-backbone atoms of the 50 ns MD simulation trajectories displayed vibrational deviations with 1.5 Å from beginning to end of simulation signifying a stable conformation of the protein and dodonic acid complex (Figure 3A, blue). While ligand RMSD was displayed very stable conformation throughout the simulation time (Figure 3A, pink). Root mean square fluctuations of the amino acid residue position of 50 ns simulation trajectories of dodonic acid bound protein was displayed in Figure 3B. The individual fluctuations of amino acid residues over a function of time from the reference structure (0 ns) after time 50 ns of the final structure of undecaprenyl diphosphate synthase displayed residue positions 160 and 170 having significant fluctuations averaging 4 Å (Figure 3B). However, no other significant fluctuating residues were observed. RMSF signify the stable conformation of the undecaprenyl diphosphate synthase and dodonic acid complex. The radius of gyration (Rg) was also determined as Rg indicates the size and compactness of the protein in the ligand-bound state. The Rg plots are displayed in Figure 3C. The Rg plot of Cα-backbone exhibited the lowering trend of trajectories throughout the simulation that signify the compactness of the undecaprenyl diphosphate synthase and dodonic acid complex. Number of hydrogen bonds analysis has great importance of studying protein ligand interaction and to understand proper binding of ligand in protein. In this study, the number of hydrogen bonds formed between the dodonic acid and undecaprenyl diphosphate synthase on an average 3 throughout the simulation time for 50 ns. The significant hydrogen bonds formed between dodonic acid and undecaprenyl diphosphate synthase indicated good biding leading to significant stability (Figure 3D). 3D interaction plot displayed the formation of three significant hydrogen bonds between dodonic acid and amino acid residues Ala76, Ser78 and Asn81 of undecaprenyl diphosphate synthase (Figure 3E) corroborated the findings of simulation hydrogen bond analysis. The total energy profile of the protein ligand complex displayed a very stable formation having total energy approximately -220 kcal/mol (Figure 3F, blue) where the major contribution of coulomb energy (Figure 3F, red) while vdW (van der Waal’s) energy contributed significant but less (Figure 3F, green) toward the stability of the complex to reach global minima.
Figure 3. Molecular dynamics and simulation of dodonic acid bound S. aureus undecaprenyl diphosphate synthase (A) RMSD curve of protein (blue), and ligand (pink), (B) RMSF plot (C) Radius of gyration (Rg), (D) Number of hydrogen bonds formed between dodonic acid bound S. aureus undecaprenyl diphosphate synthase, (E) 2D interaction plot of dodonic acid bound S. aureus undecaprenyl diphosphate synthase and (F) Energy profile of S. aureus undecaprenyl diphosphate synthase bound to dodonic acid.
Dodonic acid from D. viscosa was active against six protein targets of S. aureus such as dihydrofolate reductase (2W9S), Sortase-A (1T2P), DNA gyrase (3U2D), clumping factor A (1N67), undecaprenyl diphosphate synthase (4H8E) and dehydrosqualene synthase (2ZCO) lead to show the high affinity on the S. aureus undecaprenyl diphosphate synthase (4H8E) and might be considered its main action mechanism but it needs to be validated using in-vitro studies in the future.
ACKNOWLEDGMENTS
The authors would like to thank the National University of San Marcos, approved project with code No. A21040023 (012730-R-21) for supporting this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Ulloa-Urizar G, Aguilar-Luis MA, de Lama-Odria M del C, Camarena-Lizarzaburu J, del Valle Mendoza J. Antibacterial activity of five Peruvian medicinal plants against Pseudomonas aeruginosa. Asian Pac J Trop Biomed. 2015;5(11):928-931.
Crossref - Bussmann RW, Malca-Garcia G, Glenn A, et al. Minimum inhibitory con-centrations of medicinal plants used in Northern Peru as antibacterial remedies. J Ethnopharmacol [In-ternet]. 2010;132(1):101-108.
Crossref - Villena-Tejada M, Vera-Ferchau I, Cardona-Rivero A, et al. Use of medicinal plants for COVID-19 prevention and respiratory symptom treatment during the pandemic in Cusco, Peru: A cross-sectional survey. PLOS ONE. 2021;16(9):e0257165.
Crossref - Bussmann RW, Ashley G, Sharon D, et al. Proving that Traditional Knowledge Works: The antibacterial activity of Northern Peruvian medicinal plants. Ethnobot Res Appl. 2011;9(0):067-96.
Crossref - Abdela J. Evaluation of In Vivo Antidiarrheal Activities of Hydroalcoholic Leaf Extract of Dodonaea viscosa L.(Sapindaceae) in Swiss Albino Mice. J Evid Based Integr Med. 2019;24:2515690X19891952.
Crossref - Tong ZW, Gul H, Awais M, et al. Determination of in vivo biological activities of Dodonaea viscosa flowers against CCL4 toxicity in albino mice with bioactive compound detection. Sci Rep. 2021;11(1):13336.
Crossref - Rajamanickam V, Rajasekaran A, Anandarajagopal K, Sridharan D, Selvakumar K, Rathinaraj BS. An-ti-diarrheal activity of Dodonaea viscosa root extracts. International Journal of Pharma and Bio Sciences. 2010;1(4):182-185.
- Naicker SD, Patel M. Dodonaea viscosa var. angustifolia Inhibits Germ Tube and Biofilm Formation by C. albicans. Evid Based Complement Alternat Med. 2013;2013:261978.
Crossref - Al-Snafi PDAE. A review on Dodonaea viscosa: A potential medicinal plant. IOSR Journal of Pharmacy. 2017;07(02):10-21.
Crossref - Cao S, Brodie P, Callmander M, et al. Antiproliferative Triterpenoid Saponins of Dodonaea viscosa from the Madagascar Dry Forest. J Nat Prod. 2009;72(9):1705-1707.
Crossref - Malik MN, Haq I ul, Fatima H, et al. Bioprospecting Dodonaea viscosa Jacq.; a traditional medicinal plant for antioxidant, cytotoxic, antidiabetic and antimicrobial potential. Arabian Journal of Chemistry. 2022;15(3):103688.
Crossref - Khalil NM, Sperotto JS, Manfron MP. Antiinflammatory activity and acute toxicity of Dodonaea viscosa. Fitoterapia. 2006;77(6):478-480.
Crossref - Herrera-Calderon O, Rahman MdH, Pena-Rojas G, Andia-Ayme V. Dodonaea viscosa Jacq: A medicinal plant with cytotoxic effect on colon cancer cell line (HT-29). J Pure Appl Microbiol. 2020;14(3):1927-1934.
Crossref - Hossain MA. Biological and phytochemicals review of Omani medicinal plant Dodonaea viscosa. Journal of King Saud University – Science. 2019;31(4):1089-1094.
Crossref - Omosa LK, Amugune B, Ndunda B, et al. Antimicrobial fla-vonoids and diterpenoids from Dodonaea angustifolia. South African Journal of Botany. 2014;91:58-62.
Crossref - Wu S, Huang J, Wu Q, et al. Prevalence and characterization of Staphylococcus aureus isolated from retail vegetables in China. Front Microbiol. 2018;9:1263.
Crossref - Ghosh A, Mukerjee N, Sharma B, et al. Target Specific In-hibition of Protein Tyrosine Kinase in Conjunction With Cancer and SARS-COV-2 by Olive Nutraceuticals. Front Pharmacol. 2022;12:812565.
Crossref - Herrera-Calderon O, Chacaltana-Ramos LJ, Huayanca-Gutierrez IC, Algarni MA, Alqarni M, Batiha GES. Chemical Constituents, In Vitro Antioxidant Activity and In Silico Study on NADPH Oxidase of Allium sativum L. (Garlic) Essential Oil. Antioxidants. 2021;10:1844.
Crossref - Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2021. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2021.
- Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. Journal of Medicinal Chemistry. 2000;43(20):3714-3717.
Crossref - Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the opls force field. J Chem Theory Comput. 2010;6(5):1509-1519.
Crossref - Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225-11236.
Crossref - Martyna GJ, Klein ML, Tuckerman M. Nose-Hoover chains: The canonical ensemble via continuous dynamics. The Journal of Chemical Physics. 1992;97(4):2635.
Crossref - Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics. 1994;101(5):4177.
Crossref - Schneewind O, Missiakas D. Sortases, surface proteins and their roles in Staphylococcus aureus disease and vaccine development. Microbiol Spectr. 2019;7(1).
Crossref - Alt S, Mitchenall LA, Maxwell A, Heide L. Inhibition of DNA gyrase and DNA topoisomerase IV of Staphylococcus aureus and Escherichia coli by aminocoumarin antibiotics. J Antimicrob Chemother. 2011;66(9):2061-2069.
Crossref - Ashraf S, Cheng J, Zhao X. Clumping factor A of Staphylococcus aureus interacts with AnnexinA2 on mammary epithelial cells. Sci Rep. 2017;7:40608.
Crossref - Ragi K, Kakkassery JT, Raphael VP, Johnson R, Vidhya TK. In vitro antibacterial and in silico docking studies of two Schiff bases on Staphylococcus aureus and its target proteins. Future Journal of Pharmaceutical Sciences. 2021;7:78. https://fjps.springeropen.com/articles/10.1186/s43094-021-00225-3
- Sinko W, Wang Y, Zhu W, et al. Undecaprenyl Diphosphate Synthase Inhibitors: Antibacterial Drug Leads. J Med Chem. 2014;57(13):5693-5701.
Crossref - Khurram M, Khan MA, Hameed A, Abbas N, Qayum A, Inayat H. Antibacterial Activities of Dodonaea viscosa using Contact Bioautography Technique. Molecules. 2009;14(3):1332-1341.
Crossref - Al Bimani BMH, Hossain MA. A new antimicrobial compound from the leaves of Dodonaea viscosa for infectious diseases. Bioact Materi. 2020;5(3):602-610.
Crossref - Priya VT, Balasubramanian N, Shanmugaiah V, et al. Partially purified lead molecules from Dodonaea viscosa and their antimicrobial efficacy against infectious human pathogens. J Infect Public Health. 2021;14(12):1822-1830.
Crossref - AL-Oraimi AA, Hossain MA. In Vitro Total Flavonoids Content and Antimicrobial Capacity of Different Organic Crude Extracts of Dodonaea viscosa. J Biol Act Prod Nat. 2016;6(2):150-65.
Crossref - Rao RM, Ramakrishna K, Ashapriya M. Antibacterial activity of different crude extracts of Dodonaea viscosa. J Chem Pharm Res. 2015;7(1):350-355. www.jocpr.com
- Alomari AA, Fadlelmula AA, Harzali H. Phytochemical Study and Antimicrobial Activity of Two Medicinal Plants from Al-Baha Region. Oriental Journal of Chemistry. 2019;35(6):1782-1788.
Crossref - Kiruba Daniel SCG, Vinothini G, Subramanian N, Nehru K, Sivakumar M. Biosynthesis of Cu, ZVI, and Ag nanoparticles using Dodonaea viscosa extract for antibacterial activity against human pathogens. Journal of Nanoparticle Research. 2013;15(1).
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.