ISSN: 0973-7510
E-ISSN: 2581-690X
Coronavirus disease 2019 (COVID–19) has spread rapidly as global pandemic affecting 187 countries/ regions and emerged as worldwide health crisis. Potential antiviral drugs used for the SARS -CoV-2 in clinical treatments have side effects. However, emergency vaccines are in use but despite that increase in the coronavirus cases are alarming. Thus, it is utmost need of safer antiviral agent to treat or inhibit the viral infection. Forskolin has been reported as a possible antiviral-agent. This molecule was docked with ACE2 receptor of human which is the target for the binding of S1 unit of viral S protein of SARS-CoV- 2. In silico docking was carried out on SwissDock, PatchDock and FireDock servers. The docked ACE2 structure was further docked with the RBD of the spike protein. Forskolin is able to H-bond with the hACE2 and ACE2-forskolin fails to interact with the receptor-binding domain (RBD) of the Spike protein of SARS-CoV-2. Instead, viral RBD is repulsed by the diterpene molecule through obliteration and reciprocated binding. We report first that forskolin plays a crucial role in the inhibition of protein-protein interaction of RBD and ACE2 when docked with either of the protein.
SARS-CoV-2, S protein RBD, human ACE2 receptor, forskolin, Plectranthus barbatus, Coleus forskohlii
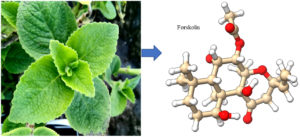
Forskolin is a small natural bioactive molecule produced by Plectranthus barbatus Andrews (syn. Coleus forskohlii Briq.) (Fig.1 A) of the plant family Lamiaceae. P. barbatus is a well-known indigenous plant of India used traditionally in various ailments since ancient times1.
Fig. 1. A – Plectranthus barbatus Plant, 1B- 3-dimensional ball and stick structure of forskolin (A major phytochemical of Plectranthus barbatus)
Forskolin (Fig1 B) is 7beta-acetoxy-8,13-epoxy-1a,6β,9a-trihydroxy-labd-14-en-11-one with the molecular formula C22H34O7. It is a lipid-soluble labdane diterpene that can penetrate cell – membranes and stimulate the enzyme adenylate cyclase2,3 which produces the ‘fight or flight’ messenger cAMP for the tissues from ATP. It is a useful chemical used in pharmaceuticals and biological research4 derived from geranylgeranyl pyrophosphate with a tetrahydropyran-derived heterocyclic ring. The topical application of forskolin has been reported to be capable of reducing intraocular pressure (IOP) in animals and human5. The molecule has been found to be useful in preventing asthma attacks in patient [6]. It is understood to work by stabilizing the cells that release histamine and other inflammatory compounds7 and by relaxing smooth muscle tissue8.
A thorough literature survey on PubMed database suggested several research articles exploring antiviral activity of plant extracts against varied coronaviruses, especially SARS-CoV9. Discovery of new lead molecules in cell-based assays (Vero E6 cells) and detection via different techniques like immune-fluorescence, western blot and flow cytometry involved analysis of roughly 10,000 compounds10. Anti-SARS-CoV activity of forskolin was reported by11 employing molecular modeling.
Here we present our observation regarding in silico docking experiments involving forskolin molecule as ligand and human cell surface protein angiotensin-converting enzyme2 (ACE2) as receptor. Spike protein of SARS-CoV-2 has receptor binding domain which specifically binds to ACE 2 (Angiotensin- converting enzyme-2) receptor. The ACE 2 receptors are expressed on the host cells12. RBD of S protein was also taken as receptor for the forskolin ligand in a separate docking experiment. Chiefly, we attempted to look into the impact of ACE2-forskolin complex on the RBD’s binding efficacy for the former which is of prime importance for successful SARS-CoV-2 infection12.
The model of SARS-CoV-2 and ACE2 complex was obtained in the expression system Spodoptera frugiperda and submitted to the Protein data bank13 with the PDB code 6vw114 contains spike protein RBD from Asn334 to Pro527and Ser19-Ala614 of ACE2 N-terminal peptidase domain. This model was downloaded, analyzed and used for studies.
Structure preparation of ligand and receptor and Molecular Docking
The 3D SDF structure (CID 47936) of forskolin molecule was retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/compound/Forskolin). UCSF chimera 1.11 [15] was used for the conversion of the SDF file into mol2 format to be used as ligand.
The 6vw1 is crystal-structure of chimeric receptor-binding domain of SARS-CoV-2 complexed with its human receptor ACE2 along with its accessory molecules, ions and water was subjected to the receptor preparation for docking. It was achieved in UCSF Chimera by removing all the nonspecific molecules and ions, chain B of ACE 2 and both chains of RBD. The prepared molecule was saved in PDB format for submission for docking16,17,18.
Docking of forskolin with ACE 2 protein chain was performed online on SwissDock web service based on EADock DSS engine19. Blind docking was performed in the vicinity of all target cavities and the binding modes with most favorable energies were evaluated with FACTS and clustered by SwissDock server. These clusters were analyzed by UCSF Chimera’s View Dock20,21 and most preferred bindings of ligand to the receptor were chosen (with lowest delta G energy). The structures were saved in pdb format for further protein-protein docking analysis.
Protein-protein Docking
ACE2 receptor with docked forskolin was subjected to protein-protein docking on PatchDock server22,23 where ACE 2 was uploaded as receptor and RBD as the protein ligand as pdb files. Best 10 solutions of the PatchDock were further uploaded to the FireDock web server for structure refinement and rescoring of protein-protein dock. The results were downloaded in pdb format and best dock structure was analyzed in UCSF Chimera.
Binding to ACE2 is indispensable for entry of SARS-CoV-2 into target cells24. The functional component in S1 subunit of S protein SARS-CoV-2 is receptor binding domain (RBD) for binding to the ACE225,26. Determination of the structure of RBD-ACE2 complex using x-ray crystallography provides insights of the residues of RBD protein that are essential for ACE2 binding. Majority of these residues are highly conserved and several of them share side chain properties with the receptor binding domain of SARS-CoV.
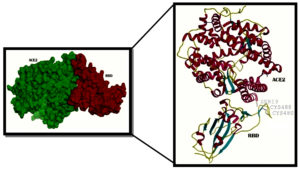
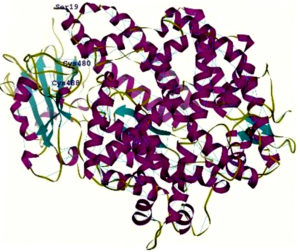
Fig. 2. A- View of the rendered complex structure of RBD (Majenta colour) and ACE2 (Green colour) B- 2 N- terminal helix of ACE2 with Ser19 interacting with the contacting segment of RBD with Cys (CYS 488 and CYS 480) residues forming a disulphide bond
6vw1 complex of RBD of SARS-CoV-2 with ACE2 shows seven antiparallel β sheets of which five make the core with short helices and loops in RBD. N terminal helix of ACE2 containing Ser19 is involved in binding to the RBD of S protein12. The peptide substrate binding site of ACE2 lies between its two lobes. Figure 2A presents the surface rendering of the complex. The segment of RBD comprising two shorter β sheets, contacts the small lobe of ACE2 to interact with many of its residues. The contacting segment of RBD has a disulfide bond between Cys480 and Cys488 in this region (Fig.2B). Properties of Ligand forskolin and ACE2 Receptor complex are presented in Table 1.
Table (1):
Properties of Ligand forskolin and ACE2 Receptor complex.
| Properties | Forskolin | Properties | SARS-CoV-2 chimeric RBD complexed with human ACE2 |
|---|---|---|---|
| PubChem ID | 47936 | PDB ID- | 6VW1 |
| Molecular Formula | C22H34O7 | Total structure weight | 192.75 kDa |
| Molecular Weight | 410.5 g/mol | Atom Count | 13180 |
| Hydrogen Bond Donor Count | 3 | Residue Count | 1579 |
| Hydrogen Bond Acceptor Count | 1 | Unique protein chains | 2 |
| Rotatable Bond Count | 3 | Co-crystalized molecules | 1.beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |
| Pharmacological properties | anti-HIV agent, a protein kinase A agonist, an adenylate cyclase agonist, an antihypertensive agent, platelet aggregation inhibitor, potent activator of adenyl cyclase system and has role in biosynthesis of cAMP, antihypertensive, positive inotropic, platelet aggregation inhibitory, and smooth muscle relaxant activities, lowers intraocular pressure, promotes release of hormones from the pituitary gland | 2. 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose 3. 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose 4. 2-acetamido-2-deoxy-beta-D-glucopyranose 5. Zinc ion 6. 1,2-ethanediol 7. Chloride ion |
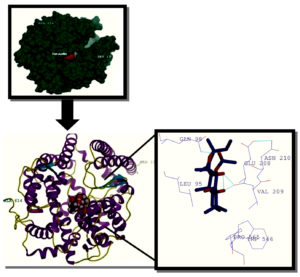
One of the best fits as derived by SwissDock for forskolin docking complex with ACE2 was Cluster 15 having Cluster rank 1. It showed Energy: 53.8005; SimpleFitness: 53.8005; FullFitness: -2634.1519; InterFull: -44.4368, IntraFull: 108.234; solvFull: -3097.68; surfFull: 399.731; extraFull: 0.0; deltaGcompsolvpol: -3097.68; deltaGcompsolvnonpol: 399.731; deltaGprotsolvpol: -3132.93; deltaGprotsolvnonpol: 401.547; deltaGligsolvpol: -10.6535; deltaGligsolvnonpol: 13.2877; deltaGvdw: -44.4368; deltaGelec: 0.0 and deltaG: -6.813238. The renderings of the complex of cluster 15 in surface and ribbon forms respectively and the ligand interaction with various residues of the receptor through H bonds and other chemical interactions at least at three positions. One such interaction is shown in Fig 3 along with the rendered surface and ribbon forms of dock structure.
Fig. 3. Cluster 15 complex rendered in UCSF chimera 10.11, Forskolin ligand molecule interactionwith receptor ACE 2- hydrogen bonds are depicted in blue colour whereas hydrophobic interaction with different aminoacid of ACE 2 protein are shown in purple colour
In PatchDock algorithm, the given molecules’ surfaces are cleaved into small patches according to shape of the surface. Similar patches are superimposed using shape matching algorithms that involve molecular shape representation and surface patch matching. Unacceptable complexes are discarded. Complexes are ranked based on its geometric shape and score of complementarities.
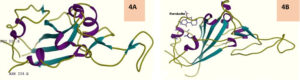
ACE2-forskolin complex and S protein RBD subjected to protein-protein docking on PatchDock and best 10 solutions uploaded to the FireDock web server gave a surprising output where Solution number 1 ranked 1 with global energy -11.19, attractive Van der Waals force (VdW) as -8.95, repulsive VdW as 2.55, Atomic contact energy (ACE) as -5.64 and hydrogen bonds contributed to the global binding energy (HB) being 00 showed only RBD to which forskolin got docked into (Fig 4A). The RBD showed an obliterated structure and obviously failed to interact with ACE2 (Fig.4B). Docking of free ACE2 with RBD on FireDock showed expected result and good intermolecular interaction (Fig.5).
Fig. 4. A Normal RBD (from 6VW1) and 4B – RBD after bound with forskolin in RBD-ACE2_Forskolin docking
Fig. 5. Protein-Protein docking of ACE2 and RBD: Without forskolin, RBD shows molecular interactions to ACE2
The above observations and inference suggest that the diterpenoid forskolin may be a good inhibitor of RBD-ACE2 binding and may prevent the entrance of SARS-CoV-2 into human cells for infection. Some related studies in literature which conceptualize on the inhibition of protein-protein binding reported compounds which binds to the region of RBD residues and thus, further impede the binding of RBD to ACE-226 where as another study involves the effect of two Chinese herbal compounds puerarin and quercetine which impair the binding of viral S protein to ACE2 receptor when docked with Human receptor for receptor binding domain of viral protein27. Only one cell line-based study involving colforsin (forskolin) where colforsin and 5 other compounds were tested and found to be broad spectrum inhibitors for spike-mediated entry of SARS-CoV-228.
We report first as per our knowledge that forskolin plays significant role in the inhibition of protein-protein interaction of SARS-CoV-2 virus receptor binding domain (RBD) of spike protein and angiotensin-converting enzyme2 (ACE2) of human lung cells through molecular docking studies. PubMed database search resulted in 427 compounds documented for covid 19 clinical trial at PubChem (https://pubchem.ncbi.nlm.nih.gov/#query=covid-19%20clinicaltrials) which don’t include Forskolin trial for SARS-CoV-2. Plant based drugs are always in high demand high efficacy, low cost, easy availability and they are used as preventive medicines29. Further studies need to be performed to analyze prophylactic properties of forskolin against SARS-CoV-2 infection but it is highly recommended by this study that use of forskolin and other natural compounds recommended by Ayurveda will play major role in keeping ourself protected from Covid 19 disease.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AK and JK conceived and executed the idea. AK and PK performed docking. AK and PK compiled the data, illustrated the Figures and tables. JK, AK and MK wrote the manuscript. JK supervised the whole work. All authors approved the final manuscript.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Kanne H, Burte NP, Prasanna V, Gujjula R. Extraction and elemental analysis of Coleus forskohlii extract. Pharmacognosy Res. 2015;7(3):237-241.
Crossref - Seamon KB, Daly JW, Metzger H, de Souza NJ, Reden J. Structure-activity relationships for activation of adenylate cyclase by the diterpene forskolin and its derivatives. J. Med. Chem.,1983;26:436-439.
Crossref - Morrone LA, Rombola L, Corasanti MT et al. Natural compounds and retinal ganglion cell neuroprotection. Progress in Brain Research. 2015;220:257-281.
Crossref - Alasbashi RH, Melzig MF. Plectranthus barbatus: A review of phytochemistry, ethnobotanical uses and pharmacology – Part I. Planta Medica. 2010;76(7):653-661.
Crossref - Caprioli J, Sears M, Bausher L et al. Forskolin lowers intralocular pressure by reducing acqueous inflow. Invest Ophthalmol Vis Sci. 1984;25(3):268-277.
- González-Sánchez R, Trujillo X, Trujillo-Hernández B, Vásquez C, Huerta M and Elizalde A. Forskolin versus sodium cromoglycate for prevention of asthma attacks: a single-blinded clinical trial. J Int Med Res. 2006;34(2):200-207.
Crossref - Marone G, Columbo M, Triggiani S et al. Forskolin inhibits the release of histamine from human basophils and mast cells. Agents Actions.1986;18:96-99.
Crossref - DeSouza NJ. Industrial development of traditional drugs: the forskolin example. A mini-review. J Ethnopharmacol.1993;38:167-175.
Crossref - Orhan EI, Deniz FSS. Natural products as potential leads against coronaviruses: Could they be encouraging structural models against SARS-CoV-2? Natural Products and Bioprospecting. 2020;10:171-186.
Crossref - , , , et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Natl. Acad. Sci. 2004;101:10012-10017.
Crossref - CC Wen, YH Kuo, JT Jan, et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem.,2007;50:4087-4095.
Crossref - Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-220.
Crossref - Helen MB, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Research., 2000;28(1):235-242.
Crossref - Shang J, Ye G, Shi K, Wan YS, Aihara H, Li F. Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2. 2020 www.rcsb.org/structure/6VW1.
Crossref - Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem., 2004;25(13):1605-1612.
Crossref - Kumar M, Topno RK, Dikhit MR, et al, Molecular docking studies of chloroquine and its derivatives against P23pro-zbd domain of chikungunya virus: Implication in designing of novel therapeutic strategies. Journal of cellular biochemistry, 2019;120(10):18298-18308.
Crossref - Kumari A, Kumar M , Sahoo GC, Kumar J. In Silico prediction of glabridin potency against human tyrosinase in hyperpigmentation condition. International Journal of Research and Analytical Reviews (IJRAR). 2021;6(2):332-341. E-ISSN 2348-1269, P- ISSN 2349-5138
- Rana S, Mahat JP, Kumar M, Sarsaiya S, Modeling and docking of cysteine protease-A (CPA) of leishmania donovani. J. Appl. Pharm. Sci.2017;7(9):179-184.
Crossref - Grosdidier A, Zoete V , Michielin O, SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Research. 2011; 39(suppl_2):W270–W277.
Crossref - Kumar M, Rana S, Kumar H, et al, Computational, structural and functional aspects of hypothetical protein of Aspergillus flavus Pheromone Receptor Pre-A (PRP-A). Journal of Applied Pharmaceutical Science. 2017;7(7):89-97.
Crossref - Kumar M, Sahoo G, Topno R, et al, Molecular interaction of antifungal compounds with fungal type I Polyketide synthase and preventing hepatocarcinogenesis. International Journal of Infectious Diseases. 2018;73:285.
Crossref - Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Efficient Unbound Docking of Rigid Molecules. In Gusfield et al., Ed. Proceedings of the 2’nd Workshop on Algorithms in Bioinformatics (WABI). Rome, Italy, Lecture Notes in Computer Science.,2002;2452: 185-200, SpringerVerlag.
Crossref - Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucl. Acids. Res. 2005;33:W363-367.
Crossref - Hoffmann M. Kleine-Weber, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.
Crossref - Walls AC, Park Y, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 ;181(2):281-292.
Crossref - Odhar HA, Ahjel SW, Hashim AF, Rayshan AM. Molecular Docking and Dynamics Simulation of a Screening Library from Life Chemicals Database for Potential Protein-Protein Interactions (PPIs) Inhibitors against SARS-CoV-2 Spike Protein. Journal of Pharmaceutical Research International. 2021;33(28):74-84.
Crossref - Pan B, Fang S, Zhang J, et al. Chinese herbal compounds against SARS-CoV-2: Puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Computational and structural biotechnology journal. 2020;18:3518-3527.
Crossref - Kumari A, J Kumar. “Phyto-Chemical Screening of Root Extracts of Glycyrrhiza Glabra by Spectroscopic Methods (UV-VIS Spectrophotometer, FTIR & HPLC)”. International Journal of Pharmaceutical Sciences and Drug Research.2019:11(6);376-381.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.