ISSN: 0973-7510
E-ISSN: 2581-690X
Allium sativum commonly known as Garlic is a familiar herb, highly studied for its valuable medicinal properties. The main objective involved in the current research is to analyze inhibitor and antibacterial action of bioactive compounds (ligands) present in the methanolic extract of Allium sativum bulbs against phytopathogen protein (receptor) through molecular docking. The effector protein AvrRps4 (4B6X) from phytopathogen Pseudomonas syringae pv. pisi, a protein responsible for Effector triggered immunity (ETI) activation and to subvert host responses in Pea plant was selected as protein target. The docking interactions between opted ligands and target protein, with ampicillin as control was done using PyRx software tool and analysed using Discovery studio 3.1 Visualiser. The outcomes obtained from in silico analysis suggested that the bioactive compound namely Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate bind effectively showing -5.7 binding energy value in comparison with antibiotic ampicillin which showed binding energy -5.8 value. This research study concluded that the bioactive compounds from methanolic extract of Allium sativum bulbs displayed a potential inhibitory activity against effector AvrRps4 protein exhibiting antibacterial properties and may be considered as possible substantial lead molecules in future prospects.
Antibacterial, Bioactive compounds, Allium sativum, Molecular docking, Effector AvrRps4 protein
Plant microbial diseases are threat to plant crops and affect good yield. Pea (Pisum sativum) is a major pulse crop grown in India, belonging to family Leguminoceae and is cultivated for dry seeds as well as green pod seeds.1 The important diseases affecting pea cultivations are caused by nematodes, viruses, bacteria and fungi. Among these, bacterial disease namely bacterial blight, a seed-borne disease, caused by Pseudomonas syringae pv. pisi, occurs to be detrimental.2
Pseudomonas syringae is one of the well-studied phytopathogen which is preferred as a model for studying plant-microbe interactions. Success of P. syringae as phytopathogen includes epiphytic survival, plant immunity quelling after infection, accession of the T3SS (type III secretion system) and an essential T3E (T3SS effectors) repository. The type III secretion system are encoded by the genes like hrp (HR and pathogenicity) and hrc (HR and conserved) genes, whereas type III secretion system effectors are encoded by avirulence (avr) and Hrp-dependent outer protein (hop) genes.3 AvrRps4 (Avirulence gene encoding resistance protein in pseudomonas syringae) is a type III secretion system effector from P. syringae pv. pisi. The amelioration and efficacy of the bacterial survival in the host metabolome is impacted by this effector protein in spite of the defense response exhibited by the host plant in order to annihilate the bacterial phytopathogen.4
Green technology involving medicinal plant extracts is an evolving method to minimize plant microbial diseases. The antibacterial and antioxidant properties of plant extracts have been reported.5 Garlic formulations are known to possess broad-spectrum antimicrobial and anti-biofilm activity on several multidrug-resistant clinically relevant bacterial species and hence could be considered as natural antibiotic agent.6 The antibacterial, antifungal, antiparasitic, antiviral, amoebicidal activities of garlic extracts have been investigated.7 Experimental evidence reported by several authors has proved the efficacy of methanolic garlic extracts and garlic essential oils against different agricultural pests. The occurrence of various phytochemicals namely saponins, steroids, terpenes, glycosides, phenols, flavonoids and alkaloids in garlic have been evaluated.8
Green technology applications in agriculture can be enhanced through Computer-aided drug discovery/design (CADD). Computational approaches help in exploring studies in molecular phytopathology by gathering information about sequences of genome, revealing 3-D structures of biomolecules etc. and play an important role in the development of antimicrobial agro-products against phytopathogens.9
Molecular docking has been shown instrumental function in the logical design of drugs.10 It is one of the techniques which indicates the favoured direction of a particle to the other and helps in forming a stable and well-constructed complex in the branch of molecular modeling.11
The current study aims to determine the phytocompounds existing in the bulb extracts of Allium sativum by OHRLCMS analysis and to analyse promising prime bioactive compounds of Allium sativum bulb extracts against chosen AvrRps4 protein by conducting molecular docking experiments.
Plant materials
Healthy, mature and fresh bulbs of Allium sativum (Bhima variety) were collected from Indian Council of Agricultural Research-Directorate of Onion and Garlic Research (ICAR – DOGR) Pune, India.
Preparation of extract
The garlic bulbs were chopped into tiny pieces, dried and powdered. About 40g of the garlic powder was weighed and extracted with 120ml of 70% methanol using Soxhlet apparatus. The Soxhlet extraction was carried out until 72 h.12 After extraction, the evaporation of the obtained solvent was done using a rotary evaporator. The obtained methanolic extract was stored at 4°C for future use.
OHR-LCMS analysis of the extracts
The High resolution Orbitrap liquid chromatography mass spectrometer (OHR-LCMS) analysis of the extract was done at Sophisticated Analytical Instrument Facility (SAIF), IIT Bombay. OHR-LCMS analysis was performed using the instrument Q-Exactive Plus Biopharma. Thermo Scientific Data Acquisition Software used for the analysis involved Thermo Scientific Xcalibur, Version 4.2.28.14 and Data Processing Software used was Compound Discoverer 2.1 SP1 and Column details included Hypersil Gold 3 micron 100 x 2.1 MM (Thermo Scientific) with Solvent A: 0.1% formic acid in Milli-Q water and Solvent B: Methanol.
Interpretation of phytocompounds
For identification of mass spectrum of OHR-LCMS, phytocompounds comparison was done with the known compounds of the spectrum using the mzCloud – Advanced Mass Spectral Database. The determination of name, molecular weight, and structure of the phytocompounds was done.13
Ligand generation
The selection of ligands were done based upon the ‘Lipinski’s rule of five’ and ADMET (Absorption, distribution, metabolism, excretion, and toxicity) studies. Structures of compounds present in A. sativum methanol bulb extract were obtained from PUBCHEM compounds database (https://pubchem.ncbi.nlm.nih.gov/). The same was applied for prediction of properties of ligands namely hydrogen donors, acceptors, logP value and molecular weight. The ligands were checked for drug-probabilities corresponding to Lipinski’s rule of five specifications namely molecular weight, log P, and many hydrogen bond donors and some hydrogen bond acceptors and the 2D structures were subject to ADMET analysis for antitoxicity properties using Data Warrior.14 The structures were replaced in SDF format and were converted into PDB format utilising PyMol.15
Retrieval and preparation of the selected protein
AvrRps4 (4B6X) protein which has a vital role in Effector triggered immunity (ETI) was targeted for its binding with the phytocompounds extracted from Allium sativum bulb methanolic extracts. The 3D structure of the AvrRps4 (4B6X) protein was obtained from Protein Data Bank (PDB) with PDB ID 4B6X_1. (https://www.rcsb.org/structure/4b6x).16
The water molecules in protein structure were removed and obtained PDB format was saved in order to get further protein validation. The structure was then improvised by allocating the bond angles, bond orders as well as topology.17 The prediction of active site pocket region was carried out using online tool CASTp.
Molecular Docking (MD) Studies
The definition of molecular docking involves “key and lock” pattern applied in order to analyse the best-possible pattern involved between receptor (ligand) and protein. Bioactive compounds revealed through OHRLCMS results of Allium sativum methanolic extract were opted for the current docking study. The appropriate bioactive chemicals chosen were docked with the target protein using PyRx software (https://sourceforge.net/projects/pyrx/). The docking interactions for the optimised ligands structural files were undertaken in order to analyse their prospects of inhibition, in opposition to the validated protein AvrRps4 (4B6X), using the software tool PyRx (https://pyrx.sourceforge.io/).18 Ampicillin was used as control.19
The genetic algorithm was applied to run the docking procedure. Depending on the RMSD (Root Mean Square Division) values the appropriate docked pattern was determined. The docked compounds were analysed using Discovery Studio 3.1 visualiser after obtaining the binding energy and the binding constants of each ligand protein complex.20 The lowest negative binding energy (∆G) specifies the strongest binding and desirable conformity for the protein and the ligand reaction.21
OHRLCMS Analysis
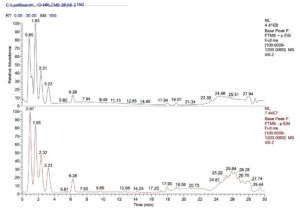
OHRLCMS evaluation was carried out for the methanolic bulb extract of Allium sativum in order to estimate its phytocompounds. The outcomes of OHRLC-MS evaluation revealed the recognition of several phytocompounds. The retention time (RT), molecular formula, molecular weight, and area (%) distribution of these phytocompounds are given in (Table 1 & 2). The OHRLCMS chromatograms of the extract are presented in Fig. 1.
Fig. 1. OHRLCMS chromatogram of methanol extract of Allium sativum bulb on positive and negative ionization mode.
OHRLCMS chromatogram (positive ionization mode) of the methanol extract of bulbs of Allium sativum indicated 17 peaks showing the appearance of seventeen compounds in total and other various compounds in minute amounts (Fig. 1). OHRLCMS chromatogram (negative ionization mode) of the methanol extract of bulbs of Allium sativum indicated 14 peaks showing the appearance of fourteen compounds in total and other various compounds in minute amounts.22 The phytocompounds in the methanolic extract of garlic bulb extract shown by OHRLC-MS of both positive ion mode and negative ion mode with their Molecular formula, Molecular weight (MW), and Retention time (RT) were given in Table 1 and Table 2 respectively.
Table (1):
Bioactive compounds identified in the methanolic extract of bulb of Allium sativum by OHRLC-MS analysis (positive ionization mode).
No. |
Compound name |
RT (min) |
Molecular wt |
Molecular formula |
Structure |
DB Difference (PPM) |
|---|---|---|---|---|---|---|
1 |
Trigonelline |
0.856 |
135.05477 |
C7H7NO2 |
99.2 |
|
2 |
Cytosine |
0.854 |
244.17897 |
C4H5N3O |
83.1 |
|
3 |
Asparagine |
0.859 |
474.16725 |
C4H8N2O3 |
93.4 |
|
4 |
4-Oxoproline |
1.636 |
129.04213 |
C5H7NO3 |
85.8 |
|
5 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
1.632 |
113.04729 |
C13H14F3NO5S |
87.6 |
|
6 |
Prolylglycine |
2.312 |
172.084 |
C7H12N2O3 |
51.4 |
|
7 |
Glycylproline |
2.317 |
580.18483 |
C7H12N2O3 |
69.1 |
|
8 |
4-Aminohippuric acid |
2.315 |
602.16664 |
C9H10N2O3 |
64.1 |
|
9 |
4-Acetamido-5-[2-(4-hydroxyphenyl)ethylamino]-5-oxopentanoic acid |
3.221 |
276.10966 |
C15H20N2O5 |
68.5 |
|
10 |
L-Phenylalanine |
3.227 |
165.07942 |
C9H11NO2 |
77.2 |
|
11 |
α-Aspartylphenylalanine |
3.227 |
588.24455 |
C13H16N2O5 |
79.6 |
|
12 |
Bromhexine |
5.602 |
110.96129 |
C14H20Br2N2 |
71.1 |
|
13 |
Dibutyl phthalate |
5.606 |
278.15245 |
C16H22O4 |
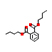 |
75.7 |
14 |
Diethyl phthalate |
5.607 |
148.01648 |
C12H14O4 |
69.4 |
Table (2):
Bioactive compounds identified in the methanolic extract of bulb of Allium sativum by OHRLC-MS analysis (negative ionization mode).
No. |
Compound name |
RT (min) |
Molecular wt |
Molecular formula |
Structure |
DB Difference (PPM) |
|---|---|---|---|---|---|---|
1 |
Melezitose |
0.973 |
323.10463 |
C18H32O16 |
74.5 |
|
2 |
N-Acetyl-1-aspartylglutamic acid |
0.976 |
682.21996 |
C11H16N2O8 |
62.9 |
|
3 |
Sucrose |
0.978 |
720.20613 |
C12H22O11 |
74 |
|
4 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
1.658 |
113.99248 |
C13H14F3NO5S |
87.6 |
|
5 |
D(-)-Amygdalin |
1.659 |
326.06962 |
C20H27NO11 |
73.5 |
|
6 |
L-Pyroglutamic acid |
2.32 |
129.04292 |
C5H7NO3 |
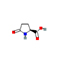 |
58.1 |
7 |
Microcystin |
2.321 |
290.09239 |
C49H74N10O12 |
63.9 |
|
8 |
Bromhexine |
2.321 |
115.04589 |
C14H20Br2N2 |
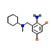 |
72 |
9 |
4-Oxoproline |
2.326 |
129.04213 |
C5H7NO3 |
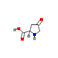 |
86.4 |
10 |
4-Oxoproline |
3.229 |
129.04213 |
C5H7NO3 |
76.9 |
|
11 |
Bromhexine |
3.248 |
114.95712 |
C14H20Br2N2 |
69.7 |
|
12 |
Bromhexine |
5.61 |
112.95665 |
C14H20Br2N2 |
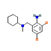 |
71.1 |
13 |
Dodecamethylcyclohexasiloxane |
5.617 |
684.19884 |
C12H36O6Si6 |
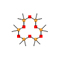 |
77.7 |
14 |
Diethyl phthalate |
5.617 |
147.08188 |
C12H14O4 |
 |
69.4 |
15 |
9-Nitrooleate |
17.202 |
545.27119 |
C18H32NO4– |
85.9 |
|
16 |
Dodecyl sulfate |
25.199 |
266.15391 |
C12H26O4S |
80.7 |
|
17 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
29.44 |
115.94092 |
C13H14F3NO5S |
60.9 |
Ligand preparation
The in silico results indicate that 5 ligands namely Dibutyl phthalate, Oxoproline, Phenylalanine, Trigonelline, Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate cleared Lipinski’s rule of five. The results include the molecular weight, log P value, number of hydrogen donors and acceptors. (Table 3). The 3D structure obtained through Insilco analysis are presented in Table 3.
Table (3):
Lipinski’s properties of the selected phytochemical compounds from bulb extracts of Allium sativum.
No |
Compound Name |
Molecular weight (<500KD) |
3D Structure |
Log P (<5) |
H-bond donor (<5) |
H-bond acceptor (<10) |
|---|---|---|---|---|---|---|
1 |
Dibutyl phthalate |
278.15 |
 |
2.26 |
1 |
2 |
2 |
Oxoproline |
129.04 |
0.51 |
2 |
3 |
|
3 |
Phenylalanine |
165.07 |
1.08 |
2 |
3 |
|
4 |
Trigonelline |
135.05 |
 |
3.11 |
0 |
2 |
5 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
115.94 |
2.32 |
3 |
2 |
Validation of the protein
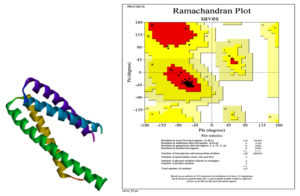
The 3D structure of the protein as a result of in silico analysis is shown in the Fig. 2.
The evaluated and validated structure of 4B6X protein was obtained using the Ramachandran plot, which predicted energetically allowed regions for backbone dihedral angles 𝜓 against 𝜙 of amino acid residues. The plots were generated by RAMPAGE server.23 The procured RAMPAGE outcomes of the proteins indicated the stability of protein structure (Fig. 2).
Docking
The antibacterial action of Allium sativum bioactive compounds in opposition to the 4B6X was performed using Pyrex. Discovery Studio 3.1 visualizer was used for analysis of the interacted complexes. The result suggests that the binding orientation differentiated according to the nature of ligand. The docking outcomes are given in Table 4 and the binding energy values obtained is summarised in Table 5.
Table (4):
The Bioactive compounds from A. sativum bulb extract docked with AvrRps4 (4B6X).
No |
Ligand |
Binding Affinity score |
No of Hydrogen Bonds |
Structure 2D |
Structure 3D |
Hydrogen Bond forming Amino Acids |
|---|---|---|---|---|---|---|
1 |
Dibutyl phthalate |
-4.5 |
1 |
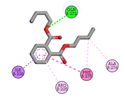 |
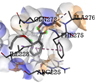 |
GLN272 |
2 |
Oxoproline |
-3.8 |
1 |
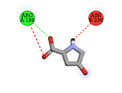 |
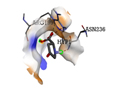 |
ARG199 |
3 |
Phenylalanine |
-4.5 |
3 |
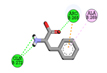 |
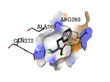 |
ARG265, GLN272, GLN272 |
4 |
Trigonelline |
-3.8 |
1 |
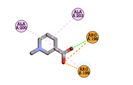 |
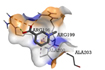 |
ARG196 |
5 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
-5.7 |
2 |
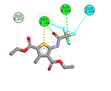 |
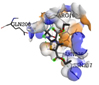 |
ASN236, ARG199 |
6 |
Ampicillin (control) |
-5.8 |
1 |
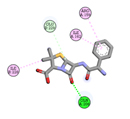 |
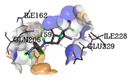 |
GLN206 |
In silico methods disclosed that opted ligands were able to interact in the active site portion of 4B6X protein and could bind in the similar hydrophobic region as seen in Ampicillin control. The outcomes of docking were expressed as e-negative values and highest negative e-values portray strongest binding affinity between the protein and ligand molecules respectively. The docked bioactive compounds revealed values differing from -3.8 to −5.7. The docking value of Ampicillin (control) in opposition to 4B6X was -5.8 kcal/mol having one hydrogen bond (Table 5).
Table (5):
Molecular docking score for Secondary metabolites of A. sativum with 4B6X.
No |
Ligand |
Docking score (kcal/mol) |
No. of hydrogen bonds formed |
|---|---|---|---|
1 |
Dibutyl phthalate |
-4.5 |
1 |
2 |
Oxoproline |
-3.8 |
1 |
3 |
Phenylalanine |
-4.5 |
3 |
4 |
Trigonelline |
-3.8 |
1 |
5 |
Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate |
-5.7 |
2 |
6 |
Ampicillin (control) |
-5.8 |
1 |
Out of the 5 ligands chosen, Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate showed maximal docking score (-5.7kcal/mol) having 2 hydrogen bonds. The next maximal docking score (-4.5 kcal/mol) containing 1 H-bond was obtained by Dibutyl phthalate followed by Trigonelline which shows the docking score of -4.5 kcal/mol. On the other hand, amino acid derivative namely phenylalanine showed the highest amount of hydrogen bonds (3 hydrogen bonds) along a docking score of -4.5kcal/mol respectively. Another amino acid derivative Oxoproline showed a docking score binding energy of -3.8 with 1 hydrogen bond. Hence, it is evident that the phytochemicals formed interaction with active site portion of the 4B6X constructively.
An untargeted quantitative OHR LC-MS phytocompounds analysis of methanolic extract of Allium sativum bulb indicated 31 total compounds. The revealed phytocompounds come under chemical groups namely saponins, glycosides, alkaloids, tannins, flavonoids, terpenes, phenol, and thiophenes.
In the drug discovery process, development in computational techniques plays a crucial role. The computational refining techniques are regularly as well as abundantly applied. The computational refining employs docking as well as scoring of every molecule in a given input and also analyses the binding patterns and interaction of one molecule (ligand) with the other (receptor) and hence helps in reducing the fare as well as duration of drug invention. Docking techniques aid to find suitable patterns of chosen ligands in the active binding pocket of a target protein and to predict their interaction in three-dimensional space.
In the present study, phytocompounds (ligands) from Allium sativum were docked with 4B6X protein to check whether these compounds can interact and circumvent the phytopathogen protein action and prove the ligand’s antibacterial effect. The out comings obtained from the in silico studies indicated that selected bioactive molecules (5 ligands) interacted with the 4B6X protein alike to Ampicillin (control). The obtained outcomes proves altogether the molecular docking method applied was evidentful in discovering naive 4B6X protein inhibitors from Allium sativum extracts.
The ligand Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate showed two hydrogen bonds with the best binding energy value in comparison with other ligands. This ligand exists in the group of thiophenes and literature study reveals that natural thiophenes are bioactive compounds and exhibit antibacterial activity antimicrobial, insecticidal and nematocidal properties under irradiation. These sulfur-containing polyacetylenes react as type II photosensitizers in presence of UV light, giving singlet oxygen that displays a wide range of microbial toxicity. The enzymatic activation of thiophenes in the root of the host plant as a result of the endoparasitic nematode attack is the substantial reason for the nematocidal activities of thiophenes. Thiophenes are considered as agricultural important agents and can act as biorational pesticides due to their photodynamic character.24
Also, other secondary metabolites which showed good docking scores namely dibutyl phthalate and trigonelline have been reported as antibacterial effective metabolites. The antibacterial activity of dibutyl phthalate has been reported.25 Trigonelline an alkaloid have antibacterial property because it can act as inhibiting ATP-dependent transport of compounds across the cell membrane, which may not allow further infection in plants.26
In summary, our in silico analysis study provides new evidence that inhibitory effect of Allium sativum bioactive compounds is promising on Pseudomonas syringae pv. pisi effector protein Avrsp4 (4B6X) that suppresses plant host immunity. From the OHRLCMS analysis of methanolic extracts of A. sativum bulb, 31 phytochemical bioactive compounds were recognised. Among these, the ligands whichever fulfilled the ‘Lipinski’s rule of five’ namely, Dibutyl phthalate, Oxoproline, Phenylalanine, Trigonelline and Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate were opted for docking. Among all the compounds, Diethyl 3-methyl-5-[(2,2,2-trifluoroacetyl)amino]thiophene-2,4-dicarboxylate showed the greatest binding affinity with least binding score of -5.7 kcal/Mol and it shows 2 hydrogen bond interactions (ASN236, ARG199) against the active site of the target protein Avrsp4 (4B6X). This result enables that A. sativum bioactive compounds could be considered for their antibacterial potential and could be a valuable source for the design and development of novel antibacterial compounds to combat bacterial plant disease process and crop damage.
ACKNOWLEDGMENTS
The authors would like to acknowledge the Management of JSS Academy of Higher Education & Research, Mysuru, Karnataka, India for supporting the basic research ideas and for the resources provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SS and KMB conceptualized the study. SS, MSM, HBG, TSG and KMB helped in manuscript preparation. SS drafted the Manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Basaiwala P, Rastogi NK, Parikh M. Genetic variability and character association in field pea (Pisum sativum L.) genotypes. The Asian Journal of Horticulture. 2013;8(1):288-291.
- Martin-Sanz A, de la Vega MP, Murillo J, Caminero C. Strains of Pseudomonas syringae pv. syringae from Pea Are Phylogenetically and Pathogenically Diverse. Phytopathology. 2013;103(7):683-681.
Crossref - Xin X-F, Kvitko B, He SY. Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol. 2018;16(5):316-328.
Crossref - Morgan K. Halane, Sang Hee Kim, Benjamin J. Spears, et al. The bacterial type III-secreted protein AvrRps4 is a bipartite effector. PLoS Pathogens. 2018 ;14(3):e1006984. Crossref
- Mansoor S, Khan I, Fatima J, Saeed M, Mustafa H. Anti-bacterial, anti-oxidant and cytotoxicity of aqueous and organic extracts of Ricinus communis. Afr J Microbiol Res. 2016;10(8):260-270.
Crossref - Zheng HM, Li HB, Wang DW, Liu D. Preparation methods for monodispersed garlic oil 872 microspheres in water using the microemulsion technique and their potential as antimicrobials. J Food Sci. 2013;78(8):1301-1306.
Crossref - Torpol K, Wiriyacharee P, Sriwattana S, Sangsuwan J, Prinyawiwatkul W. Antimicrobia 832 activity of garlic (Allium sativum L.) and holy basil (Ocimum sanctum L.) essential oils applied by liquid vs. vapour phases. Int J Food Sci Technol. 2018;53(9):2119-2128.
Crossref - Ali M, Ibrahim IS. Phytochemical Screening and Proximate Analysis of Garlic (Allium Sativum). Arc Org Inorg Chem Sci. 2019; 4(1):478-482.
Crossref - Shanmugam G, Jeon J. Computer-Aided Drug Discovery in Plant Pathology. Plant Pathol J. 2017;33(6):529-542.
Crossref - Zammel N, Saeed M, Bouali N, et al. Antioxidant and Anti-Inflammatory Effects of Zingiber officinale roscoe and Allium subhirsutum: In Silico. Biochemical and Histological Study. Foods. 2021;10(6):1383.
Crossref - Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019;20(18):4331.
Crossref - Eidi A, Eidi M, Esmaeili E. Antidiabetic effect of garlic (Allium sativum L.) in normal and streptozotocin-induced diabetic rats. Phytomedicine. 2006;13(9-10):624-629.
Crossref - Fabresse N, Larabi IA, Stratton T, et al. Development of a sensitive untargeted LC-HRMS screening devoted to hair analysis through a shared MS2 spectra database: a step towards early detection of new psychoactive substances. Drug Testing and Analysis. 2019;11(5):697-708.
Crossref - Shivakumar R, Venkatarangaiah K, Shastri S, Nagaraja RB, Sheshagiri A. Antibacterial property and molecular docking studies of leaf calli phytochemicals of Bridelia scandens Wild. Pharmacog J. 2018;10(6):1221-1229.
Crossref - Lill MA, Danielson ML. Computer-aided drug design platform using PyMOL. J Comput Aided Mol Des. 2011;25:13-19.
Crossref - Sohn KH, Hughes RK, Piquerez SJ, Jones JDG, Banfield MJ. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci U S A.. 2012;109(40):16371-16376.
Crossref - Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotoninO-methyltransferase using different docking routines. T Bio Med. 2013;10(1):63.
Crossref - Trott O, Olson AJ. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461.
Crossref - Shukla A, Khan E, Srivastava A, Tandon P, Sinha K. A computational study on molecular structure,multiple interactions, chemical reactivity and molecular docking studies on 6[D (−) α-aminophenyl- acetamido] penicillanic acid (ampicillin). Molecular Simulation. 2016;42(11):863-873.
Crossref - Kemmish H, Fasnacht M, Yan L. Fully automated antibody structure prediction using BIOVIA tools: validation study. PLoS One. 2017;12(5):e0177923.
Crossref - Afriza D, Suriyah WH, Ichwan SJ. In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. J Phys Conf Ser. 2018;1073(3):1-7.
Crossref - Ramakrishnan P, Kalakandan SK, Pakkirisamy M. Studies on Positive and Negative ionization mode of ESI-LC-MS/MS for screening of Phytochemicals on Cassia auriculata (Aavaram Poo). Pharmacognosy Journal. 2018;10(3):457-462.
Crossref - Ho BK, Brasseur R. The Ramachandran Plots of Glycine and Pre-Proline. BMC Structural Biology. 2005;5:14.
Crossref - Prasad KC, Angothu BN, Latha TM, Nagulu M. Synthesis of some novel 2 aminothiophene derivatives and evaluation for their antimicrobial activity. Int J Pharm Bio Sci. 2017;7(1):01-07.
Crossref - Shobi TM, Viswanathan MBG. Antibacterial activity of di-butyl phthalate isolated from Begonia malabarica. J Appl Biotechnol Bioeng. 2018;5(2):97-100.
Crossref - Tariverdizadeh N, Mohebodini M, Chamani E, Ebadi A. Iron and zinc oxide nanoparticles: An efficient elicitor to enhance trigonelline alkaloid production in hairy roots of fenugreek. Industrial Crops & Products. 2021;162:113240.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.