ISSN: 0973-7510
E-ISSN: 2581-690X
Escherichia coli (E. coli) is the predominant pathogen causing urinary tract infection. E. coli expresses a wide spectrum of virulence factors that enable it to cause the disease. The present study was conducted on 50 E. coli isolates to assess some important phenotypic virulence factors of E. coli: haemolytic activity, cytotoxicity and haemagglutination and their relations to the following virulence genes: hlyA, cnf-1 and papC. Fluoroquinolones susceptibility was also assessed. Fifty E. coli isolates were isolated from non-hospitalized patients with UTIs and identified by colony morphology and standard biochemical reactions. The isolates were examined phenotypically for haemolytic activity using 5% sheep blood agar, cytotoxicity by their cultivation on HEp-2 cell line and haemagglutination of group O human RBCs. Virulence genes were detected by multiplex PCR. Fluoroquinolones susceptibility was done using disk diffusion method. Phenotypic methods showed that 30% of the isolates were b- haemolytic, 18% of the isolates showed haemagglutination with human RBCs, with higher prevalence for MRHA (12%) than MSHA (6%), and 92% of the isolates showed morphological changes, detachment and lysis in HEp-2 cells. Multiplex PCR showed that the most prevalent gene was cnf-1 (30%), while 26% of isolates were positive for hlyA and 26% of isolates were positive for papC. Only one UPEC isolate showed the simultaneous presence of papC, cnf-1 and hlyA, while 10 isolates were harbouring hlyA and cnf-1, 2 isolates were harbouring cnf-1 and papC genes and 2 isolates were harbouring hlyA and papC simultaneously. Fluoroquinolones susceptibility showed that 60% of the isolates were resistant to the used fluoroquinolones.
Escherichia coli, virulence, multiplex PCR, Fluoroquinolones susceptibility
Urinary tract infection (UTI) is among the most common bacterial infectious diseases encountered at all ages. Escherichia coli is being the etiologic agent in 50-80%. (Farell et al., 2003; Matute et al., 2004; Zhanel et al., 2006). Certain strains of E. coli are consistently associated with uropathogenicity and are designated as uropathogenic E. coli (UPEC) (Connell et al., 1996). UPEC strains encode widespread virulence factors closely related to colonization, persistence, and pathogenesis of bacteria in the urinary tract; the most important of these factors include adhesins or fimbriae, biofilm formation, siderophores and toxins such as haemolysins (Hly) (Dhakal et al., 2008) and cytotoxic necrotizing factor-1 (CNF-1) (Wiles et al., 2008a). The virulence factors among UTI agents are really different and clinical isolates could not be detected as UPEC by using traditional methods. Therefore, using molecular detection methods is critical for identification of UPEC strains (Saraylu et al., 2012). In clinical management, it is usually accepted that resistance, especially multidrug resistance, equates to the virulence of the strain. However, molecular studies have indicated an inverse relationship between the distribution of virulence factors and antimicrobial resistance, particularly to quinolones as well as fluoroquinolones (Vila et al., 2002; Johnson et al., 2003; Moreno et al., 2006; Piatti et al., 2008).
Aim of Work
The present study was conducted to detect some important virulence factors of E. coli phenotypically: haemolytic activity, cytotoxic activity and haemagglutination and related virulence genes: hlyA, cnf-1 and papC, by using multiplex PCR. Study the relation between the bacterial resistance to fluoroquinolones and the virulence potential of E. coli.
The study has been approved by the Research and Ethical Committee of Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University.
Isolation and Identification of E. coli
Urine samples from non-hospitalized cases of urinary tract infections were collected from clinical laboratories of Kasr El Ainy hospital. Microscopic examination of a wet film of urine was carried out. Samples with pus cells ³ 10/ HPF were quantitatively inoculated on MacConkey’s agar (Oxoid, UK) and then incubated at 37 ºC for 24 hrs. Culture plates with bacterial count ³ 105 CFU/ ml and growth of single morphotype of lactose fermenting colonies were selected for identification of E. coli by standard biochemical methods.
Phenotypic Detection of UPEC Virulence Factors
The E. coli isolates were tested for expression of haemolytic, cytotoxic activity and haemagglutination as described below:
Assessment of haemolytic activity
The cytolytic toxin secreted by most haemolytic E. coli isolates is known as alpha haemolysin. E. coli isolates were subcultured onto 5% sheep blood agar (Oxoid, UK) and incubated overnight at 37ºC. Haemolysin production was detected by presence of a zone of complete haemolysis around the inoculation site (Raksha et al., 2003; EL-Mosallamy et al., 2015; Tabasi et al., 2015).
Assessment of cytotoxic activity
The cytotoxic effect of E. coli isolates was detected according to Ghadir et al. (2010), however, HEp-2 cells was used instead of Vero cells according to Mills et al. (2000).
Preparation of bacterial isolates
E. coli isolates were obtained from frozen stock and subcultured on blood agar (Oxoid, UK) then on MacConkey’s agar (Oxoid, UK). Colonies were then inoculated in test tubes containing 10 ml of tryptic soy broth (TSB) (Oxoid, UK) and incubated for 18-20 h at 37ºC and kept at 4ºC and used for inoculum preparation. Bacteria were harvested by centrifugation at 3000 rpm (MSE Centaur 2, UK). The supernatant was discarded and the bacterial pellets were washed once with phosphate buffered saline (PBS), then resuspended in Eagle’s minimum essential medium with Earl’s balanced salts (EMEM) (Lonza, Belgium) and adjusted to a count1×108 CFU/ ml.
Cell line and growth conditions
HEp-2 cells were maintained in EMEM (Lonza, Belgium) supplemented with 2% fetal bovine serum (FBS) (Lonza, Belgium), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in 5 % CO2 and subcultured every 3-4 days. HEp-2 cells suspended in MEM Earl’s and supplemented with 10 % FBS were seeded in 24-well tissue culture plates and kept at 37°C with 5 % CO2 for 24 hours to form a confluent monolayer. 100 µl of the bacterial cell suspension prepared above was added to each well and incubated at 37ºC in 5 % CO2 for 3 hours. HEp-2 cell morphology was assessed by examination using an inverted microscope (Olympus bx 50, Japan) 1, 2 and 3 hours post inoculation.
Assessment of haemagglutination
E. coli isolates were inoculated into nutrient broth and incubated at 37 °C for 48 hours for production of fimbriae. Five milliliter of group O venous blood was collected then washed three times in normal saline and a suspension of 3% RBCs in phosphate buffered saline (pH 7.4) was prepared (Lonza, Belgium). They were used immediately or within a week when stored at 3-5 °C. The slide haemagglutination test was carried out on a multiple-concavity slide by adding 50 µl of the RBC suspension to 50 µl of the broth culture and the slide was rocked at room temperature for 5 minutes. Presence of clumping was taken as positive for haemagglutination. At the same time, 50 µl of 3% D-mannose (Fluka AG, Switzerland) was added to the red cells and broth culture in another set and rocked as above. Haemagglutination inhibited in presence of D-mannose was labeled as mannose sensitive haemagglutination (MSHA); indicating type1 fimbriae, if agglutination occurred even in presence of D-mannose, it was called mannose resistant haemagglutination (MRHA); indicating presence of P fimbriae (Uma et al., 2013).
Molecular Detection of UPEC Virulence Genes
E. coli isolates were tested for the presence of virulence genes including: hlyA, cnf-1and papC using multiplex PCR.
DNA Extraction
DNA extraction was performed for all the clinical isolates using CinnaPure DNA extraction kit (Tehran, Iran) according to manufacturer’s instructions.
Amplification
Amplification was done according to Bashir et al. (2012) using 50 µl reaction mixture for each isolate. Primers sequences are illustrated in Table (1).
Table (1):
Primers’ Sequences.
| Primer | Sequence | Reference |
|---|---|---|
| hly A | F: 5ˊAACAAGGATAAGCACTGTTCTGGCT3ˊ | Yamamoto et al. (1995) |
| R: 5ˊACCATATAAGCGGTCATTCCCGTCA3ˊ | ||
| cnf-1 | F: 5ˊAAGATGGAGTTTCCTATGCAGGAG3ˊ | Arisoy et al. (2006) |
| R: 5ˊCATTCAGAGTCCTGCCCTCATTAT3ˊ | ||
| papC | F: 5ˊGACGGCTGTACTGCAGGGTGTGGCG3ˊ | Daigle et al. (1994) |
| R: 5ˊATATCCTTTCTGCAGGGATGCAATA3ˊ |
Table (2):
Expected PCR product size.
Genes |
Amplicon size(bp) |
Reference |
|---|---|---|
hly A |
1177 |
Yamamoto et al. (1995) |
cnf-1 |
498 |
Arisoy et al. (2006) |
papC |
328 |
Daigle et al. (1994) |
The thermal cycler (Biometra T 3000) conditions were as follows: an initial denaturation step at 94°C for 5 min followed by 30 cycles of amplification (denaturation at 94°C for 1 min, annealing at 56°C for 1 min and extension at 72°C for 1 min) and final extension of 5 min at 72°C.
Agarose Gel Electrophoresis
Amplified PCR products were visualized by 2% (w/v) agarose gel electrophoresis (stained with ethidium bromide; 0.5 µg/ml) at 120 volts for 45 minutes. The bands were visualized under UV transilluminator (Biometra Ti 3) and photographed. 100 bp DNA ladder H3 RTU (GeneDire, Taiwan) (which gave 12 discrete bands at 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1500 and 3000 bp) was used.
Fluoroquinolones susceptibility using Kirby Bauer Disk Diffusion method
Disk diffusion method was done using fluoroquinolones: ciprofloxacin (CIP 5µg), levofloxacin (LEV 5µg), norfloxacin (NOR 10µg), and ofloxacin (OFX 5µg) (Oxoid, UK).
The E. coli colonies were suspended in sterile normal saline and adjusted to 0.5 MacFarland standard, then inoculated on Muller-Hinton agar plates and incubated with the previously mentioned antimicrobial disks at 35ºC ±2 for 18 hours. The interpretation of the diameters of the inhibition zones were done according to CLSI, (2014) (Table 3).
Table (3):
Interpretation of sensitivity testing of E. coli with fluoroquinolones disks.
| Strain | Antimicrobial agent | Disk content | Zone diameter | ||
|---|---|---|---|---|---|
| R | I | S | |||
| E. coli | Ciprofloxacin | 5 µg | ≤15 | 16-20 | ≥21 |
| Levofloxacin | 5 µg | ≤13 | 14-16 | ≥17 | |
| Norfloxacin | 10 µg | ≤12 | 13-16 | ≥17 | |
| Ofloxacin | 5 µg | ≤12 | 13-15 | ≥16 | |
Table (4):
Distribution of Virulence genes among E. coli isolates.
No. of Virulence genes |
Virulence genes |
No. of isolates |
|---|---|---|
3 |
cnf-1, hlyA, papC |
1 |
2 |
cnf-1, hlyA |
10 |
2 |
cnf-1, papC |
2 |
2 |
hlyA, papC |
2 |
1 |
cnf-1 |
2 |
1 |
papC |
8 |
Total |
25 |
Statistical Analysis
Data were statistically described in terms of frequencies (number of cases) and percentages and compared using Chi-square (c2) test. Exact test was used instead when the expected frequency is less than 5, P values less than 0.05 was considered statistically significant. All statistical calculations were done using computer program SPSS (Statistical Package for the Social Science; SPSS Inc., Chicago, IL, USA) release 15 for Microsoft Windows.
The present study was performed on 50 E. coli isolates isolated from urine samples of patients with UTI. E. coli isolates were identified by their colony morphology on MacConkey’s agar and standard biochemical reactions.
Phenotypic Detection of Virulence Factors of E. coli Isolates
Haemolytic activity
In the current study 15 (30%) isolates showed b-haemolysis, while the remaining 35 (70%) isolates were non-haemolytic.
Cytotoxic assay
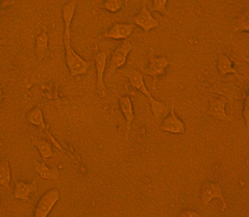
Upon visual assessment of the HEp-2 cell line one hour after infection with 1 x 108 CFU/ ml of the tested isolates, no effect was seen on HEp-2 cells. Two hours post infection; it was found that out of 50 UPEC isolates, 46 (92%) isolates showed cytotoxic effect with mild detachment and lysis of HEp-2 cell line and almost complete detachment of the cell line 3 hours post infection, while only 4 (8%) isolates showed no effect on the cell line (Figures 1, 2 and 3).
Fig. 2. The cytotoxic effect of the tested E. coli isolate on HEp-2 cells with mild detachment 2 h post infection
Fig. 3. The cytotoxic effect of the tested E. coli isolates on HEp-2 cells with almost complete detachment 3 h post-infection
Haemagglutination assay
Out of the 50 E. coli isolates, 9 (18%) isolates showed haemagglutination (HA) of group O RBCs, while 41 (82%) were non-haemagglutinating. Out of the 9 isolates positive for HA, 6 isolates were MRHA (6/50, 12%) and 3 isolates were MSHA (3/50, 6%).
Molecular Detection of E. coli Virulence Genes
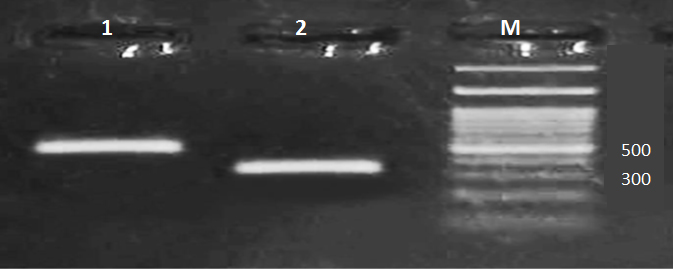
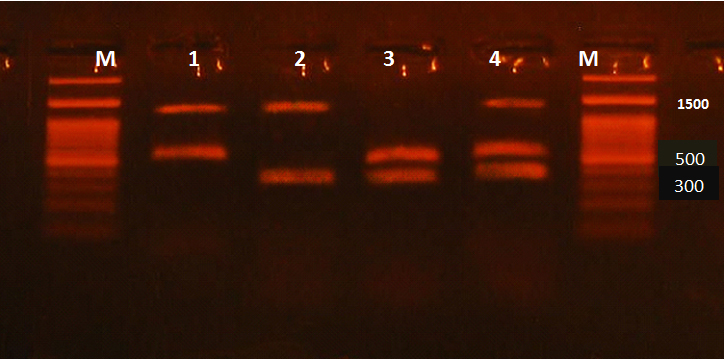
Virulence genes including hlyA, cnf-1 and papC of 50 UPEC were detected by the multiplex PCR. Amplicon sizes of hlyA, cnf-1 and papC were 1177, 498, 328 (bp), respectively (Figure 4 and 5).
Fig. 4. Multiplex PCR for virulence genes in E. coli isolates. Lane (1): cnf-1 (band at 498 bp), Lane (2): papC (band at 328 bp), Lane (M): 100 bp DNA ladder
Fig. 5. Multiplex PCR for virulence genes in E. coli isolates. Lane (1): hlyA and cnf-1, Lane (2): hlyA and papC, Lane (3): cnf-1 and papC, Lane (4): hlyA, cnf-1 and papC, Lane (M): 100 bp DNA ladder
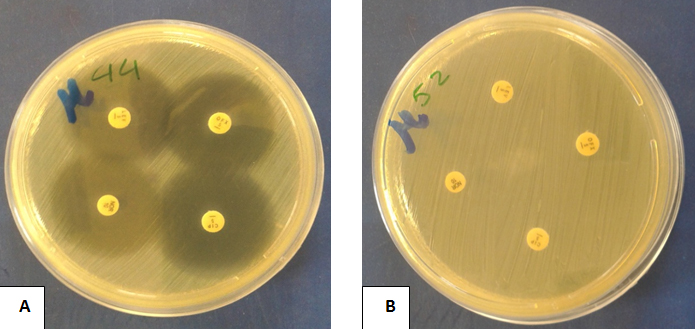
Fig. 6. Fluoroquinolones susceptibility of isolated E. coli to different fluoroquinolones using disk diffusion method (A): An E. coli isolate showing sensitivity to all used fuoroquinolones (B): An E. coli isolate showing resistance to all used fuoroquinolones
Twenty five isolates were positive for one or more of the targeted 3 genes, while the remaining isolates were negative for the targeted genes. It was found that one isolate was positive for the three targeted virulence genes, 10 isolates showed the presence of both cnf-1 and hlyA, 2 isolates showed the presence of both cnf-1 and papC, 2 isolates showed the presence of both hlyA and papC, while 8 isolates showed the presence of papC and 2 isolates showed the presence of cnf-1 gene (Table 4).
It was found that the most prevalent gene was cnf-1 which was detected in 15 (30%) isolates, while 13 (26%) isolates were positive for hlyA and 13 (26%) isolates were positive for papC.
Virulence Factors in Relation to Virulence Genes
Haemolytic activity in relation to hlyA gene
It was found that hlyA gene was detected in higher percentage in b- haemolytic isolates (61.5%) than non- haemolytic isolates (38.5%) with statistically significant difference (P- value = 0 .011).
Cytotoxicity in relation to cnf-1 and hlyA
Out of 46 isolates showing cytotoxicity by phenotypic methods, 17 (37%) isolates were positive for the cytotoxic genes; 11 isolates were positive for both cnf-1 and hlyA gene, while 4 isolates were only positive for cnf-1 gene and 2 isolates were only positive for hlyA gene. The 4 cytotoxicity negative isolates expressed neither of the 2 genes.
Haemagglutination (HA) in relation to papC gene
Out of the 9 isolates showing haemagglutination by phenotypic methods; only 3 (33%) isolates were positive for papC gene (2 isolates were MRHA and one isolate was MSHA). On the other hand, 10 (24.4%) out of the 41 non-haemagglutinating isolates were positive for papC gene. Although, haemagglutination positive isolates showed a higher percentage of papC gene than the non- haemagglutinating isolates there was no statistically significant difference between them (P- value = 0.679).
Results of Fluoroquinolones Sensitivity of the E. coli Isolates
In vitro susceptibility pattern of E. coli isolates showed that 30 (60%) isolates of E. coli were resistant to all fluoroquinolones, while 20 (40%) isolates were sensitive (Figure 6).
Association of Virulence Factors Expression with Resistance to Fluoroquinolones among E. coli Isolates
Haemolytic activity in relation to fluoroquinolones resistance
Phenotypic analysis showed that out of the 15 isolates with b- haemolysis; 10 (33.3%) were fluoroquinolone resistant and 5 (25%) were fluoroquinolones sensitive isolates with no statistically significant difference between the two groups (P- value = 0.380).
Cytotoxicity in relation to fluoroquinolones resistance
Out of the 46 isolates showing cytotoxicity to HEp-2 cells; 27 (90%) isolates were fluoroquinolone resistant and 19 (95%) isolates were fluoroquinolones sensitive with no statistically significant difference between the two groups (P- value = 0.641).
Haemagglutination in relation to fluoroquinolones resistance
Out of the 9 haemagglutinating isolates; 5 (16.6%) isolates were fluoroquinolones resistant, (3 MRHA and 2 MSHA), while 4 (20%) isolates were fluoroquinolones sensitive, (3 MRHA and 1 MSHA), with no statistically significant difference between the two groups (P- value = 0.456).
Association of Virulence Genes with Resistance to Fluoroquinolones among E. coli Isolates
Out of the 30 fluoroquinolones resistant isolates; 16 (53.3%) isolates harboured the virulence genes with variable distribution. It was found that hlyA, cnf-1 and papC genes were detected in the following percentages: 27%, 30% and 30%, respectively. On the other hand, in the susceptible group 8 (40%) isolates harboured the virulence genes with variable distribution. It was found that hlyA, cnf-1 and papC genes were detected in the following percentages respectively 25%, 30% and 20%. There was no statistically significant difference between the resistant and susceptible groups regarding the 3 virulence genes.
UPEC is the most frequent cause of community and hospital-acquired urinary tract infections (Calhau et al., 2015). These infections can vary from a simple cystitis to a serious parenchymatous attack. The degree of severity depends on the virulence of the responsible isolates (Srinivasan et al., 2003; Soto et al., 2011).
Several virulence factors such as fimbriae, toxins, and siderophores contribute to the colonization and pathogenicity of UPEC (Qin et al., 2013). Measuring a phenotype in vitro does not always correlate with in vivo expression and very often underestimates the presence of a virulence factor in vivo (Kaper et al., 2004). The distribution of virulence properties can vary depending upon host characters, type of infection and predisposing factors which determine the host parasite interaction in vivo which can culminate in an active infection (Vijayalakshmi et al., 2015). In addition there is always the possibility of mutation at the level of the corresponding gene, leading to the absence of its detection. Therefore, a positive PCR shows the presence of the virulence gene, but a negative PCR does not point to the absence of the corresponding operon (Tarchouna et al., 2013).
This study was designed to assess some important phenotypic virulence factors of E. coli: haemolytic activity, cytotoxic activity and haemagglutination and their relations to the following genes: hlyA, cnf-1 and papC.
We found that 30 % of E. coli isolates showed b- haemolysis on sheep blood agar. Similar results were obtained by other studies done by Fatima et al. (2012), EL-Mosallamy et al. (2015), Tabasi et al. (2015) and Vijayalakshmi et al. (2015) who reported that 30%, 24%, 34% and 25% of the isolates were b- haemolytic, respectively.
Higher rates of b- haemolytic E. coli were detected by Mandal et al. (2001), Raksha et al. (2003), Rebecca and Elizabeth (2005), Santo et al. (2006), Regua- Mangia et al. (2010) and Sai et al. (2013) who reported that 45.5%, 41.36%, 40.7%, 96%, 47 % and 40% of E. coli isolates were b- haemolytic respectively. Several reasons explain the differences in the percentage of haemolysin production by UPEC like: source of blood, type of haemolysin produced, source of bacteria and method to screen the production ability (Al-Chalabi et al., 2010).
Regarding cytotoxic effects of E. coli in this study, it was found that no change was detected in the HEp-2 cell line during the first hour of its incubation with E. coli suspension. However, 2 hours post infection; 92% of the tested E. coli isolates showed morphological changes and almost complete detachment in HEp-2 cells was detected 3 hours post infection. Our results were in accordance with Guignot et al. (2000) and Ghadir et al. (2010).
Agglutination of human erythrocytes by E. coli strain is an indirect evidence of the presence of fimbriae on that strain. MRHA of human RBCs is the phenotypic expression of P fimbriae on E. coli, while MSHA is the phenotypic expression of type 1 fimbriae and both of them mediate adherence and they are important in bladder colonization (Vijayalakshmi et al., 2015).
In the present study, it was found that 18% of the isolates showed haemagglutination. Similar results were obtained by Vidhya et al. (2016) who found that 20% of the isolates were positive for haemagglutination. Higher rates were reported by Fatima et al. (2012), Desai et al. (2013), Sai et al. (2013) and Vijayalakshmi et al. (2015) who found that 48%, 66%, 59% and 68% of the isolates were haemagglutinating, respectively.
In this study, higher prevalence for MRHA (12%) than MSHA (6%) was observed. Our results were in agreement with Fatima et al. (2012) and Desai et al. (2013). On the other hand, higher prevalence for MSHA than MRHA was recorded by Sai et al. (2013) and Vijayalakshmi et al. (2015).
The genes coding for alpha-haemolysin (hlyA), cytotoxic necrotizing factor (cnf-1) and P fimbrial adhesin (papC) were detected by PCR in the current study. It was observed that 13 isolates (26%) of E. coli isolates were positive for hlyA gene which was in agreement with Bingen-Bidois et al. (2002), Tiba et al. (2008), Bashir et al. (2012) and Fatima et al. (2012). Lower rates were obtained by other studies done by Arisoy et al. (2006), Farshad and Emamghorashi (2009) and Tarchouna et al. (2013). Higher rates were obtained by Santo et al. (2006) and Karimian et al. (2012) who reported that 96% and 50.4% of the isolates were positive for the gene, respectively.
As regard the cnf-1 gene, it was found that 15 isolates (30%) of E. coli isolates in this study were positive for the gene. Similar results were obtained by Adwan et al. (2015) and Calhau et al. (2015). Lower results were obtained by Arisoy et al. (2006), and Tarchouna et al. (2013). On the other hand, higher rates were obtained by Karimian et al. (2012) who reported that 50.4% of the isolates were positive for the gene.
In the present study, it was found that 13 isolates (26%) were positive for papC gene. This finding was in accordance with Farshad and Emamghorashi (2009), Qin et al. (2013) and Calhau et al. (2015). Higher papC gene carriage rates of 36%, 32%, 32.7% and 34.6% were reported by Usein et al.(2001), Santo et al. (2006), Tiba et al. (2008), Firoozeh et al. (2014), respectively. Lower papC gene carriage rates of 23% and 24% were obtained by Arisoy et al. (2006) and Bashir et al. (2012), respectively. The diversity in frequency of pap gene among different studies can be attributed to the fact that UPEC strains can utilize a variety of adhesins to bind to the urinary epithelial cells, and start the infection. Hence, the strains lacking the pap operon may use other adhesins encoding operons such as afa, and sfa for binding (Neamati et al., 2015).
In addition, the differences in prevalence of UPEC virulence genes showed that the virulence properties of UPEC strains is closely depending on geographic region, weather climate of each regions, diets, the levels of public health, hospital’s health and even methods of sampling (Karimian et al., 2012).
In the present study, the simultaneous presence of papC, cnf-1 and hlyA was observed in only one UPEC isolate. Bashir et al. (2012) found that the three genes were found collectively in 4 isolates (out of a total of 59 isolates). The cumulative presence of papC, cnf-1 and hlyA is the evidence of the presence of pathogenicity island IIJ96, which is highly prevalent among UTI isolates (Piatti et al., 2008).
Regarding the relation between the phenotypic virulence factors and the virulence genes, it was found that hlyA gene was detected in a higher percentage in b-haemolytic E. coli 53% (8/13) than non- haemolytic E. coli 14% (5/13) and this difference was statistically significant (P- value = 0.011). The presence of the gene in some non-haemolytic strains may be explained by the possibility of modification of the gene leading to inactive haemolysin (Landraud et al., 2000). Also, since alpha haemolysin is a pore-forming toxin, at low concentration its binding does not always lead to lysis (Boehm et al., 1990; Ostolaza and Goni, 1995). On the other hand, 7 b-haemolytic E. coli isolates in this study were negative for hlyA gene which means that haemolysis on sheep blood agar might be due to haemolysins other than a-haemolysin (Fatima et al., 2012).
In our study, we found that all isolates positive for hlyA gene exhibited cytotoxic activity on HEp-2 cell line; 11 of which harboured cnf-1 gene in addition. Out of the 13 isolates positive for papC gene, only 2 isolates were positive for MRHA. This can be explained by Wullt (2002) who found that some papC positive isolates, especially those isolated form asymptomatic infections, do not express P fimbriae. In addition, the expression of P fimbriae is subjected to phase variation so in vitro growth conditions differing in glucose concentration, temperature and pH may switch the fimbriae on or off. We found that the remaining 4 isolates positive for MRHA were negative for papC gene. According to Sabitha et al. (2014), MRHA can be mediated by P fimbriae and also by X, F1C and Dr fimbriae.
Regarding sensitivity of E. coli to fluoroquinolones, it was observed that 30 isolates (60%) were resistant to all tested fluoroquinolones. This result was in agreement with Lüpez-Banda et al. (2014), Calhau et al. (2015) and Gururaju et al. (2015).
In the current study, although there was a higher percentage of both virulence factors and virulence genes in the fluoroquinolones resistant group than in the fluoroquinolones sensitive group, this difference was not statistically significant. This was in accordance with Oliveira et al. (2011). According to previous studies, Johnson et al. (2003), Horcajada et al. (2005), Moreno et al. (2006), Takahashi et al. (2009), and Kawamura-Sato et al. (2010) found that quinolones and fluoroquinolones resistant UPEC strains display overall reduced virulence while, susceptible E. coli strains were more virulent. Piatti et al. (2008) found a lower incidence of papC, hlyA, and cnf-1 among fluoroquinolones resistant group than among susceptible group, which seems to be due to a loss of the corresponding PAI, probably as a result of the mutation that causes resistance.
Virulence determinants may help us to get insights into pathogenesis of UPEC isolates and management of UTIs. Phenotypic methods are simple but not very specific, while multiplex PCR is rapid, easy and more specific in detecting virulence genes. The tested E. coli isolates exhibited a high resistance to fluoroquinolones with no significant difference was found between fluoroquinolones resistant and sensitive E. coli isolates regarding the virulence genes as well as virulence factors.
ACKNOWLEDGMENTS
This study was funded partly by Faculty of Medicine, Cairo University and partly by the authors.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Adwan G., Issa B. and Adwan K. Virulence profile, fluoroquinolone and quinolone resistance of uropathogenic Escherichia coli isolates recovered from Thabet Hospital-Tulkarm, Palestine. Br. Microbiol. Res. J., 2015; 5(5):412-423.
- Al-Chalabi R., Al-Ubaidy A. and Al-Ibadi M. Detection of urovirulence genes (eae, E-hly, á-hly) of uropathogenic Escherichia coli by specific PCR.J. Biotechnol. Res. Center, 2010; 4(1): 44-54.
- Arisoy M., Aysev D., Ekim M., et al. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int. J. Clin. Pract., 2006; 60: 170-173.
- Bashir S., Haque A., Sarwar Y., et al. Virulence profile of different phylogenetic groups of locally isolated community acquired uropathogenic E. coli from Faisalabad region of Pakistan. Ann. Clin. Microbiol. Antimicrob., 2012; 11 (23): 1-6.
- Bingen-Bidois M., Clermont O., Bonacorsi S., et al: Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun., 2002; 70: 216-226.
- Boehm D., Welch R. and Snyder I. : Domains of Escherichia coli haemolysin (HlyA) involved in binding of calcium and erythrocyte membranes. Infect. Immun., 1990; 58: 1959-1964.
- Calhau V., Domingues S., Ribeiro G., et al. Interplay between pathogenicity island carriage, resistance profile and plasmid acquisition in uropathogenic Escherichia coli. J. Med. Microbiol., 2015; 64: 828-835.
- Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing, twenty-fourth informational supplement, document M100-S24. CLSI, Wayne, Pa, USA 2014.
- Connell I., Agace W., Klemm P., et al.: Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci., 1996; 93(18): 9827-9832.
- Daigle F., Harel J., Fairbrother J., et al. Expression and detection of pap, sfa, and afa-encoded fimbrial adhesion systems among uropathogenic Escherichia coli. Clin. J. Microbiol., 1994; 40: 286-291.
- Desai S., Rajput A., Kagal A., et al: Virulence factors in uropathogenic Escherichia coli (upec) causing urinary tract infections. Ind. J. Basic and Applied Med. Res., 2013; 2: 886-896.
- Dhakal B., Kulesus R. and Mulvey M. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur. J. Clin. Invest., 2008; 38 (2): 2-11.
- EL-Mosallamy W., Desouky S., Abo El-Azm A., et al. Detection of some virulence factors and pyelonephritis-associated pilus (pap) encoding operon gene in uropathogenic Escherichia coli. Egy. J. Med. Microbiol., 2015; 24(3): 37-43.
- Farell D., Morrissay I., De Rubeids D., et al. A UK multicentre study and the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J. Infect., 2003; 46: 94-100.
- Farshad S. and Emamghorashi F. The prevalence of virulence genes of E. coli strains isolated from children with urinary tract infection. Saudi J. Kidney Dis. Transpl., 2009; 20(4): 613-617.
- Fatima N., Agrawal M., Shukla I., et al. Characterization of uropathogenic E. coli in relation to virulence factors. Open Access Sci. Rep., 2012; 1 (7): 1-4.
- Firoozeh F., Saffari M., Neamati F., et al. Detection of virulence genes in Escherichia coli isolated from patients with cystitis and pyelonephritis. Int. J. Infect. Dis. 2014; 29: 219-222.
- Ghadir S., Aboulwafa M. and Hassouna N. Cytotoxic activities of some Escherichia coli isolates: Possible mechanisms and approaches for inhibition. J. Am. Sci., 2010; 6: 269-283.
- Guignot J., Breard J., Bernet-Camard M., et al. Pyelonephritogenic diffusely adhering Escherichia coli EC7372 harboring Dr-II adhesin carries classical uropathogenic virulence genes and promotes cell lysis and apoptosis in polarized epithelial CaCo-2/Tc7 cells. Infect. Immun., 2000; 68 (12): 7018-7027.
- Gururaju T., Kasturi T. and Mallikarjuna Reddy C. A study of antibiotic sensitivity pattern and detection of fluoroquinolones resistance to Escherichia coli from urinary tract infections. Int. J. Curr. Microbiol. App. Sci., 2015; 4(2): 799-805.
- Horcajada J., Soto S., Gajewski A., et al. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J. Clin. Microbiol., 2005; 43: 2962-2964.
- Johnson J., Kuskowski M., Owens K., et al. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J. Infect. Dis., 2003; 188: 759-768.
- Kaper J., Nataro J. and Mobley H. (2004): Pathogenic Escherichia coli. Nat. Rev. Microbiol., 2 (2): 123-140.
- Karimian A., Momtaz H. and Madani M. Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr. J.Microbiol. Res., 2012; 6(39): 6811-6816.
- Kawamura-Sato K., Yoshida R., Shibayama K., et al. Virulence genes, quinolone and fluoroquinolone resistance, and phylogenetic background of uropathogenic Escherichia coli strains isolated in Japan. Jpn. J. Infect. Dis., 2010; 63: 113-115.
- Landraud L., Gauthier M., Fosse T., et al. Frequency of Escherichia coli strains producing the cytotoxic necrotizing factor (CNF-1) in nosocomial urinary tract infections. Lett. App. Microbiol., 2000; 30(3): 213-216.
- López-Banda D., Carrillo-Casas E., Leyva-Leyva M., et al. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Bio. Med. Res. Int., 2014; 1-10.
- Mandal P., Kapil A., Goswami K., et al. Uropathogenic Escherichia coli causing urinary tract infections. Indian J. Med. Res., 2001; 114: 207-211.
- Matute A., Hake E. and Schurink C. Resistance of uropathogens in symptomatic urinary tract infection in Leon, Nicaragua. Int. J. Antimicrob. Agents, 2004; 25: 506-509.
- Mills M., Meysick K. and O’Brien A. Cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli kills cultured human uroepithelial 5637 cells by an apoptotic mechanism. Infec. Immun., 2000; 68(10): 5869-5880.
- Moreno E., Prats G., Sabaté M., et al. Quinolone, fluoroquinolone and trimethoprim/ sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J. Antimicrob. Chemo., 2006; 57: 204-211.
- Neamati F., Firoozeh F., Saffari M., et al. Virulence genes and antimicrobial resistance pattern in uropathogenic Escherichia coli isolated from hospitalized patients in Kashan. Iran. Jundishapur J. Microbiol., 2015; 8(2):1-6.
- Oliveira F., Paludo K., Arend L., et al. Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genetics and Molecular Research; 2011; 10(4): 4114-4125.
- Ostolaza H. and Goni F. Interaction of the bacterial protein toxin alpha-haemolysin with model membranes: protein binding does not always lead to lytic activity. FEBS Lett., 1995; 371: 303-306.
- Piatti G., Mannini A., Balistreri M., et al. Virulence factors in urinary Escherichia coli strains: Phylogenetic background and quinolone and fluoroquinolone resistance. J. Clin. Microbiol., 2008 ; 46: 480-487.
- Qin X., Hu F., Wu S., et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS One, 2013; 8(4): 1-7.
- Raksha R., Srinivasa H. and Macaden R. Occurrence and characterization of uropathogenic Escherichia coli in urinary tract infections. Ind. J. Med. Microbiol., 2003 ;21: 102-107.
- Rebecca N. and Elizabeth M. Some virulence characteristics of uropathogenic Escherichia coli in different patient groups. Indian J. Med. Res., 2005; 122: 143-147.
- Regua-Mangia A., Irino K., Pacheco R., et al. Molecular characterization of uropathogenic and diarrheagenic E. coli pathotypes.J. Basic Microbiol., 2010; 50: 107-115.
- Sabitha B., Vimal K. and Geetha R. Determination of phenotypic expression of the fimbriae and haemolysin of uropathogenic Escherichia coli (UPEC). Int. J. Adv. Med., 2014; 1(1):13-18.
- Sai S., Hema P. and Sreenivasa R. Virulence-associated factors in Escherichia coli strains isolated from urinary tract infections. Int. J. Curr. Microbiol. App. Sci., 2013; 2(10): 436-440.
- Santo E., Macedo C. and Marin J. Virulence factors of uropathogenic Escherichia coli from a University Hospital in Ribeirão Preto, São Paulo, Brazil. Rev. Inst. Med. trop., 2006; 48(4): 185-188.
- Saraylu J., Mehrabadi J., Fooladi A., et al.: Prevalence and evaluation of toxin genes among uropathogenic Escherichia coli clinical isolates by duplex PCR. J. Med. Bacteriol., 2012; 1(1):17-22.
- Soto S., Zuniga S., Ulleryd P., et al.: Acquisition of pathogenicity island in an Escherichia coli clinical isolate causing febrile urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis., 2011; 10: 1258-1262.
- Srinivasan U., Foxman B. and Marrs C. Identification of a gene encoding heat resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J. Clin. Microbiol., 2003; 41: 285-289.
- Tabasi M., Karam M., Habibi M., et al: Phenotypic assays to determine virulence factors of uropathogenic Escherichia coli (UPEC) isolates and their correlation with antibiotic resistance pattern. Osong Public Health Res. Perspect, 2015; 6 (4): 261-268.
- Takahashi A., Muratani T., Yasuda M., et al. Genetic profile of flouroquinolone- resistant E. coli isolated from cystitis: phylogeny, virulence factors, PIS usp– subtype and mutation pattern. J. Clin. Microbiol., 2009; 49: 791-795.
- Tarchouna M., Ferjani A., Ben-Selma W., et al. Distribution of uropathogenic virulence genes in Escherichia coliisolated from patients with urinary tract infection. Int. J. Infect. Dis., 2013; 17: 450-453.
- Tiba M., Yano T. and Leite D. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev. Inst. Med. trop., 2008; 50 (5): 255-260.
- Uma B., Sunitha P., Pattepu R., et al. Hemagglutination and biofilm formation as virulence markers of uropathogenic Escherichia coli in acute urinary tract infections and urolithiasis. Ind. J. Urol., 2013; 29 (4): 277-281.
- Usein C., Damian M., Tatu-Chitoiu D., et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J. Cell. Mol. Med., 2001; 5: 303-310.
- Vidhya J., Samuel S., Kennedy K., et al. Phenotyphic determination of urinary virulence factors in Escherichia coli. Inter. J. Recent Trends in Sci. Tech., 2016; 19(1): 13-16.
- Vijayalakshmi J., Hymavathi R., Renuka Devi A., et al. Study of virulence factors in uropathogenic Escherichia coli. J. Evol. Med. Dent. Sci., 2015; 4(8): 1297-1305.
- Vila J., Simon K., Ruiz J., et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J. Infect. Dis., 2002; 186: 1039-1042.
- Wiles T., Dhakal B., Eto D., et al : Inactivation of host AKT/PKB signaling by bacterial pore-forming toxins. Mol. Biol. Cell. 2008a; 19(4): 1427-1438.
- Wullt B. The role of P fimbriae for Escherichia coli establishment and mucosal inflammation in the human urinary tract. Int. J. Antimicrob. Agents, 2002; 21: 605-621.
- Yamamoto S., Terai A., Yuri K., et al. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol., 1995; 12: 85-90.
- Zhanel G., Hisanaga T. and Laing N. Antibiotic resistance in Escherichia coli outpatient urinary isolates: final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA). Int. J. Antimicrob. Agents, 2006; 27: 468-475.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.