ISSN: 0973-7510
E-ISSN: 2581-690X
Contaminated surfaces increase the risk of hospital infections. Traditional hospital cleanliness monitoring has become insufficient. ATP bioluminescence is a developed monitoring tool with limited clinical data in healthcare settings. Therefore, the current work aims to study the impact of the ATP monitoring tool on wound infection rates and fecal colonization among burn patients. The study was designed over two phases. Phase I involved conventional cleaning monitoring by visual inspection, while phase II involved the ATP bioluminescence tool. In both phases, clinical and environmental swabs were collected for microbial culture and identification. Gram-negative bacteria were screened for carbapenem resistance. Among the five selected cases, MALDI-TOF and Vitek2 were utilized to test for phenotypic relatedness between common isolates from different clinical and environmental sources. The wound infection rate was significantly reduced from 23% in phase I to 8% in phase II (p-value <0.005). Fecal colonization by CR bacteria demonstrated 7% and 14% in phase I and phase II, respectively. Environmental culture demonstrated significantly decreased microbial isolation rates from 37% (phase I) to 10% (phase II) (p-value<0.001) with a non-significant decrease in CR bacteria. Total pass and failed cleaning rates for ATP bioluminescence were 70.9% and 6.08%, respectively. Common isolates in 3 cases exhibited a similarity of >65% by MALDI-TOF and the identical resistance phenotypes by Vitek2. The ATP bioluminescence cleaning verification system has been proven a rapid and objective tool that positively impacts microbial isolation rates from clinical and environmental samples.
ATP Monitoring, Infections, CRE, Burn, MALDI-TOF
Wound infection is a significant global concern because it accounts for roughly one-quarter of all healthcare-associated infections (HAI) and fatalities in burn patients,1 who are vulnerable to acquiring infections due to poor skin integrity.2 Contaminated surfaces and inadequate cleaning of the hospital environment significantly contribute to the occurrence of HAI and fecal colonization by superbugs, including carbapenem-resistant (CR) organisms, with adverse consequences of prolonged hospitalization as well as increased morbidity and mortality.3 This urges healthcare facilities to adopt monitoring tools to ensure adequate environmental cleaning.4
Visual inspection and microbiological culture are conventional methods for evaluating surface cleaning. Visual inspection is simple, inexpensive, and easy to perform; however, it is not objective in assessing adequate cleaning. Microbiological culture provides information on surface microbial contaminants, but it is time-consuming. With the evidence of the inadequacy of conventional methods, alternative methods have been developed to improve the monitoring of cleaning.5 Adenosine triphosphate (ATP) bioluminescence assay provides immediate feedback on the quality of cleaning.6 ATP is a substance present in living cells and organic materials. The presence of ATP on a surface indicates either microbial contamination or other organic material.7 The ATP monitoring system can determine the number of organic matter residuals that remains after cleaning an environmental surface, surgical instrument, or medical device.8 ATP bioluminescence can be employed systematically and regularly in training and education to provide immediate feedback to environmental service workers or as a quality indicator for cleaning.9 This technology is more popular in the food and pharmaceutical industry; nevertheless, there is a scarcity of data on its clinical use in healthcare facilities.9
Consequently, the present work aims to investigate the impact of using ATP bioluminescence as a monitoring tool for environmental cleaning on rates of nosocomial wound infections and rectal colonization among burn patients in a tertiary-care hospital. In addition, it aims to characterize clinical and environmental isolated organisms and study the phenotypic relatedness between common isolates.
Our study was conducted at the burn unit of Kasr Al-Ainy tertiary care hospital over two equal phases (6 months), separated by a run period (2 months). The study describes the rates of patient rectal colonization and wound infection with the characterized isolated microorganisms. The study was carried out with the conventional monitoring of environment cleaning (visual observation) in phase I in comparison with the ATP bioluminescence monitoring tool in phase II.
Study Design
During both phases, clinical samples in the form of wound swabs were collected from 100 burn patients 48 hours after hospital admission in order to assess hospital-acquired infections, and rectal swabs were taken on the day of admission (R0) and four days later (R4) in order to assess hospital-acquired colonization. Environmental swabs were collected from bed frames in the burn unit, as depicted in Table 1. The two-month run period between both phases was specified for introducing the ATP bioluminescence, training on its working procedure, and establishing cut-off pass and fail relative light units (RLU) limits for different environmental surfaces.
Table (1):
Work design and time frame of the study.
| Study period (14 months) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | M12 | M13 | M14 |
| Phase I | Run period | Phase II | |||||||||||
| • R0: rectal swab day 0 • R4: rectal swab day 4 • Wound swab • Environmental swab (bed frame) |
Training on the newly introduced ATP bioluminescence and setting up RLU limits | • R0: rectal swab day 0 • R4: rectal swab day 4 • Wound swab • Environmental swab (bed frame) • ATP bioluminescence swabs (bed frames, nurse desk, tray table, light switch) |
|||||||||||
Clinical and Environmental Sampling
Rectal swabs were collected from patients on day zero (R0) and day four (R4) of admission to the burn unit. A cotton swab was placed into the rectum and gently rotated.10 After washing with saline, wound swabs were rotated over a 1 cm2 area of the wound.11 Environmental swabs were collected from bed frames using moistened sterile cotton swabs over a 10 cm2 area.12 For environmental cleaning, a prepared 0.1% active chlorine solution (1000 ppm) was utilized.13
ATP Bioluminescence Surface Cleaning Verification
The Hygiena™ SystemSURE Plus™ cleaning verification system (Hygiena, LLC, US) was utilized for measuring the level of ATP present as a surrogate for surface microbial contamination with Bioluminescence Technology UltraSnap. The UltraSnap contains a luciferase enzyme, which converts ATP to adenosine monophosphate and produces light quantified in RLU by the SystemSURE Plus™ luminometer device.
During the run period, pass and fail limits of ATP were set for each of the high touch tested surfaces (bed frame, tray, nurse desk, and light switch) as guided by the manufacturer. At each location, 5-10 replicate ATP tests were conducted. The pass limit was determined by calculating the average RLU of the 5-10 test results, whereas the fail limit was determined by multiplying the pass limit by 3. The caution range was the range between the pass and fail limits.
In phase II, ATP swabs were collected from high-touch surfaces. As described by the manufacturer, swabbing was conducted by rotating the swab over an area of 10×10 cm, which was inserted into the device. ATP results were displayed on the digital screen within 15 seconds and were interpreted as recommended by the manufacturer: a) pass: successful cleaning, no action required; b) fail: inadequate cleaning, recleaning required; c) caution: the surface may not have been adequately cleaned.
Microbial Isolation and Identification
All clinical and environmental swabs in both phases were sent to the microbiology laboratory, where they were streaked on Blood and MacConkey culture media before incubation at 37°C for 24 hrs. Standard microbiological procedures were used to identify the isolated organisms.14 CR Gram-negative organisms, including Enterobacteriaceae (CRE), were screened by testing susceptibility to meropenem (10 mcg, Oxoid) by adopting the Kirby Bauer disc diffusion method.15
Phenotyping by MALDI-TOF and Vitek2
Concomitantly recovered organisms of the same type from clinical and environmental samples were tested for their phenotypic relatedness by MALDI-TOF (Matrix-Assisted Laser Desorption Ionization-Time of Flight) and automated Vitek2 advanced expert system (AES). MALDI-TOF was performed following the manufacturer’s protocol. Microbial proteins were extracted, then ionized using laser, and separated according to mass/charge ratio. Cluster analysis of proteomic profiles was created utilizing MALDI Biotyper 3.1 (Bruker Daltonics, Bremen, Germany) database version 3.3.1. The manufacturer defined the similarity index between the same species as 65%. Vitek2 (Biomerieux, France) is an automated turbidimetric system for microbial identification and detection of resistance profiles.16
Statistical Analysis
Data were analyzed using the Statistical Package for Social Science (IBM SPSS) version 23. The comparison between groups was carried out using Chi-square test (p-value > 0.05: non-significant; p-value < 0.05: significant).
In the present study, clinical samples were collected in the form of rectal (R0 and R4) and wound swabs from two groups of burn patients: phase I (n=100) and phase II (n=100). In each phase, environmental swabs (n=100) were collected from patients’ bed frames.
Clinical and Environmental Microbial Isolates
For rectal swabs, a total of 107 and 106 microbial isolates were grown from R0 and 44 and 36 isolates from R4, in phases I and II, respectively. As shown in Table 2, E. coli and Klebsiella were the most frequently recovered Gram-negative organisms, while Enterococci and S. aureus was the predominant Gram-positive organisms in both phases. Only the reduced recovery of Enterococci from 5 isolates in phase I compared to no recovery in phase II was statistically significant (p-value: 0.03).
Table (2):
Distribution of microbial isolates among clinical and environmental samples.
| Types of isolates | Clinical and environmental isolates N (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R0 | R4 | Wound | E | |||||||||
| P I N=107 |
P II N=106 |
P-value | P I N=44 |
P II N=36 |
P-value | P I N=23 |
P II N=8 |
P- value |
P I N=37 |
P II N=10 |
P-value | |
| E. coli | 43 40.2% |
40 37.7% |
0.713 | 17 38.6% |
18 50.0% |
0.308 | 2 8.7% | 2 25.0% |
0.236 | 12 32.4% |
4 40.0% |
0.654 |
| Klebsiella | 27 25.2% |
28 26.4% |
0.843 | 11 25.0% |
13 36.1% |
0.281 | 4 17.4% |
1 12.5% |
0.746 | 6 16.2% |
4 40.0% |
0.103 |
| Pseudomonas | 1 0.9% |
3 2.8% |
0.308 | 2 4.5% |
0 0.0% |
0.195 | 11 47.8% |
3 37.0% |
0.613 | 4 10.8% |
0 0.0% |
0.277 |
| Acinetobacter | 1 0.9% |
3 2.8% |
0.308 | 1 2.3% |
0 0.0% |
0.363 | 2 8.7% |
0 0.0% |
0.388 | 3 8.1% |
0 0.0% |
0.352 |
| Proteus | 1 0.9% |
1 0.9% |
1.000 | 0 0.0% |
0 0.0% |
1.000 | 0 0.0% |
0 0.0% |
1.000 | 3 8.1% |
0 0.0% |
0.352 |
| S. aureus | 14 13.1% |
6 5.7% |
0.063 | 3 6.8% |
4 11.1% |
0.499 | 0 0.0% |
1 (12.5%) |
0.085 | 6 16.2% |
1 10.0% |
0.624 |
| Enterococci | 16 15.0% |
17 16.0% |
0.827 | 5 11.4% |
0 0.0% |
0.037 | 1 4.3% |
0 (0.0%) |
0.549 | 1 2.7% |
0 0.0% |
0.599 |
| Nonhemolytic Streptococcus | 2 1.9% |
2 1.9% |
1.000 | 1 2.3% |
0 0.0% |
0.363 | 0 0.0% |
1 (12.5%) |
0.085 | 2 5.4% |
0 0.0% |
0.452 |
| Methicillin-Resistant S. aureus | 0 0.0% |
0 0.0% |
1.000 | 0 0.0% |
0 0.0% |
1.000 | 3 13.0% |
0 (0.0%) |
0.283 | 0 0.0% |
0 0.0% |
1.000 |
| Candida | 2 1.9% |
6 5.7% |
0.146 | 4 9.1% |
1 2.8% |
0.246 | 0 0.0% |
0 (0.0%) |
1.000 | 0 0.0% |
1 5.4% |
0.052 |
R0: rectal isolates at day 0 of admission; R4: rectal isolates at day 4 of admission; W: wound; E: environmental isolates; P I: phase I; P II: phase II; P-value: < 0.05: significant, > 0.05: non-significant
In wound cultures, the infection rate was significantly reduced from 23% in phase I to 8% in phase II (P<0.05). Pseudomonas and Klebsiella were the most common Gram-negative bacteria in phase I isolates, accounting for 47.8 % and 17.4 %, respectively. Pseudomonas and Klebsiella frequencies were reduced in phase II compared to phase I, with no statistically significant difference in Acinetobacter, MRSA, or Enterococci growth.
In phase I, environmental swab cultures revealed a microbial contamination rate of 37%, which was reduced to 10% in phase II (P<0.0001). As displayed in Table 2, E. coli and Klebsiella were predominant in phase I with reduced frequencies in phase II. Phase II cultures did not show Pseudomonas, Acinetobacter, Enterococci, and Streptococci growth, with no statistically significant difference compared to phase I.
CR Gram-negative isolates were found in clinical and environmental samples at rates of 19% and 15%, respectively, in phases I and II (p-value: 0.57). Rectal colonization by CR bacteria occurred at total rates of 7% and 14% in phases I and II, respectively (p-value: 0.16), as depicted in Table 3. In wound culture, CR bacteria were detected at a rate of 8% in phase I with a significant reduction to a single grown CR Pseudomonas isolate (1%) in phase II (p-value <0.05). However, no CRE was recovered in both phases. Environmental cultures recovered 4 CR Gram-negative bacteria in phase I but none in phase II, with no significant difference (p-value: 0.12).
Table (3):
Distribution of carbapenem-resistant bacteria in both phases.
| Phase I | Phase II | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R0 | R4 | W | E | Total | R0 | R4 | W | E | Total | |
| CR E. coli | 2 | 2 | 0 | 0 | 4 | 3 | 2 | 0 | 0 | 5 |
| CR Klebsiella | 1 | 0 | 0 | 0 | 1 | 3 | 3 | 0 | 0 | 6 |
| Total CRE | 3 | 2 | 0 | 0 | 5 | 6 | 5 | 0 | 0 | 11 |
| CR Pseudomonas | 0 | 0 | 6 | 3 | 9 | 0 | 0 | 1 | 0 | 1 |
| CR Acinetobacter | 1 | 1 | 2 | 1 | 5 | 3 | 0 | 0 | 0 | 3 |
| Total CR bacteria | 4 | 3 | 8 | 4 | 19 | 9 | 5 | 1 | 0 | 15 |
CR: carbapenem-resistant, and CRE: carbapenem-resistant Enterobacteriaceae (CR E. coli + CR Klebsiella).
ATP Bioluminescence in Monitoring Environment Cleaning
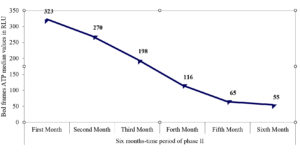
A total of 148 ATP swabs were collected from high-touch surfaces. Table 4 illustrates the interpretative results for the ATP swabs according to the preset cut-off range for every surface. Total pass and fail results were recorded with rates of 70.9% and 6.08%, respectively, and detailed in Table 4. The median and interquartile ranges (IQR) for ATP amounts on the tray, nurse desk, and light switch swabs were 85 (39–183), 219 (314–901), and 133.5 (56–183), respectively. As shown in Figure, there was a decline in ATP amount over the six months of phase II for bed frames, with recorded median values (IQR) of 323 (175.5–507), 270 (183–405), 197.5 (116.5–327.5), 116 (66–147), 65 (47–183), and 55 (33–153), respectively.
Table (4):
Interpretative ATP results for environmental swabs from high-touch surfaces.
Interpretative results |
Bed (n=112) |
Light (n=10) |
Tray (n= 13) |
Desk (n=13) |
Total (n=148) |
|---|---|---|---|---|---|
Cut-off ranges |
266-798 |
148-445 |
115–344 |
250-900 |
|
Pass N (%) |
78 (69.6%) |
10 (100%) |
9 (69.2%) |
8 (61.5%) |
105 (70.9%) |
Caution N (%) |
30 (26.7%) |
– |
4 (30.8%) |
– |
34 (22.9%) |
Fail N (%) |
4 (3.57%) |
– |
– |
5 (38.5%) |
9 (6.08%) |
Phenotyping by MALDI-TOF and Vitek2 System
The concomitant recovery of the same type of bacterial isolates from different clinical and environmental sources occurred at rates of 13% in phase I and 11% in phase II. As shown in Table 5, the highest frequency occurred for recovery of common isolates from R0 + E swabs at rates of 6% and 4% in phase I and phase II, respectively. Five cases were selected to test for microbial relatedness by MALDI-TOF and Vitek2 AES system. As depicted in Table 6, MALDI-TOF recorded a similarity of >65% among common Klebsiella isolates from R4+E in cases 1 and from R0+E in case 2 as well as among common E. coli isolates from R0+E in case 3 with the identical resistance phenotypes detected by Vitek2.
Table (5):
The frequency of common recovery of the same type of isolates from clinical and environmental samples in phase I compared to phase II.
| Source of recovery | Phase I patients (n=100) |
Phase II patients (n=100) |
P-value | ||
|---|---|---|---|---|---|
| Type of organism | Frequency N (%) |
Type of organism | Frequency N (%) |
||
| R0 + R4 + wound | Klebsiella | 3 (3%) |
E. coli | 1 (1%) | 0.308 |
| R0 + R4 + E | – | 0 (0%) |
Klebsiella E. coli |
2 2 (4%) |
0.277 |
| R0 + E | E. coli | 6 (6%) |
E. coli Klebsiella |
3 1 (4%) |
0.757 |
| R4 + E | Klebsiella E. coli |
2 1 (3%) |
Klebsiella | 1(1%) | 0.308 |
| Wound + E | Pseudomonas | 1 (1%) |
– | 0 (0%) |
0.316 |
| R0 + wound | – | 0 (0%) | Klebsiella | 1 (1%) |
0.316 |
| Total | 13 (13%) | 11 (11%) | 0.66 | ||
Table (6):
Phenotyping results of common recovered isolates by MALDI-TOF and Vitek2 in 5 selected events.
| Events | Common isolates from different sources | |||||||
|---|---|---|---|---|---|---|---|---|
| Source of recovery | Types of isolates | Similarity index by MALDI-TOF | Resistance phenotypes by Vitek2 | |||||
| Beta-lactams | Carbapenems | Quinolones | SXT | Aminoglycosides | ||||
| Case 1 | R4 + E | Klebsiella pneumoniae | >65% | Resistant: ESBL | Susceptible | Susceptible | Resistant | Resistant |
| Case 2 | R0 + E | Klebsiella pneumoniae | >65% | Resistant: ESBL | Susceptible | Susceptible | Resistant | Resistant |
| Case 3 | R0 + E | E. coli | >65% | Resistant: acquired penicillinase | Resistant | Susceptible | Resistant | Susceptible |
| Case 4 | R0 + R4 + E | E. coli | <65% | Resistant: acquired penicillinase (R0, R4) Resistant: ESBL (E) |
Resistant (R0, R4) Susceptible (E) |
Susceptible (R0, R4) Resistant (E) |
Resistant (R0, R4) Resistant (E) |
Susceptible (R0, R4) Resistant (E) |
| Case 5 | R0 + E | E. coli | <65% | Resistant: acquired penicillinase | Susceptible | Susceptible | Resistant | Susceptible |
AES: Advanced Expert System of Vitek2 system, ESBL: extended spectrum beta-lactamases, and SXT: trimethoprim-sulfamethoxazole.
In the present study, wound infection occurred with a rate of 23% in phase I, and a reduction to 8% was observed in phase II, indicating that ATP bioluminescence may significantly contribute to improving cleaning effectiveness. The majority of isolates were Gram-negative organisms, consistent with another study conducted in a burn ICU.17 Pseudomonas was the predominant isolate, accounting for 47.8% in phase I and 37% in phase II. This finding agrees with several studies that reported Pseudomonas spp. as the predominant bacteria in burn wounds, ranging from 20% to 40%.18–20 In contrast, other studies found that S. aureus was the predominant bacterial isolate in burn wounds with reported rates of 47.6%, 33.8%, and 22.4%, respectively.21-23 The present study revealed wound infection by CR Gram-negative organisms at rates of 8% in phase I and 1% in phase II, which may highlight the beneficial role of ATP bioluminescence tool in reducing the possibilities of transmitting infection. No CRE were isolated from wounds in both phases, unlike other reports of higher recovery of CRE from wounds with rates of 0.3–2.4%.23 In our study, rectal colonization by CRE was found at rates of 5% in phase I and 11% in phase II, where all cases were found to have CRE colonization on admission. Our rates were lower than other reported rates (18.9%–45%) by several other studies.24–26 This could be explained by different study populations and healthcare settings, as CRE colonization is usually more reported in ICU settings.25 The unexpectedly higher CRE fecal colonization rate in phase II than in phase I could be attributed to more cases already being admitted with CRE fecal colonization than in phase I.
Environmental culture revealed a predominance of Gram-negative organisms showing a rate of 28% in phase I, which was reduced to 8% in phase II, with E. coli and Klebsiella being the most frequently isolated organisms with rates of 16% and 10%, respectively. This finding is consistent with the study of Huang et al., who reported that Gram-negative bacteria were predominantly isolated from high touch surfaces at rates of 18.8% and 8.2% before and after cleaning, respectively.9 The predominance of E. coli and Klebsiella in the current study agrees with universal reports on common nosocomial pathogens.27 In contrast, other studies demonstrated the dominance of Staphylococci isolated from environmental surfaces.28 Variation in microbial patterns among different studies might be related to different study populations, geographical distribution, and different pathogens inhabiting healthcare centers.29
In our study, only four CR organisms were isolated from environmental surfaces in phase I with absent recovery in phase II, denoting the role of ATP bioluminescence in improving the cleaning practices and reducing surface contamination by superbugs. This finding aligns with the study of Peter et al., who reported reduced isolation of MRSA in enhanced cleaning monitoring compared to the standard cleaning observation.30 Indeed, no environmental contamination by CRE was recorded in our study, in contrast to other reported rates of contamination that reached 8.4%, which can be explained by variable infection control policies and levels of compliance among different healthcare settings.31,32
In our study, the recorded events of concomitant recovery of clinical and/or environmental common isolates and the detected species similarity by MALDI-TOF strongly suggest that hospital surfaces are usually critical factors in microbial circulation. By ATP monitoring, reduced events in phase II may support improved cleaning effectiveness. ATP bioluminescence revealed that 70.9 percent of collected swabs were successfully cleaned, while 6.08 % were inadequately cleaned. The highest rates of pass results were observed for the light switch (100%) and bed frames (69.6%). Bed swabs revealed a gradual decline in ATP amounts over the six months of phase II, which may reflect improved cleaning effectiveness due to the corrective actions taken to respond to failed results. This finding strongly agrees with previous reports on a significant decline in mean values of ATP on bed frames after versus before cleaning.33,34
Conventional methods of evaluating cleanliness have become insufficient. Due to its speed and objectivity, ATP bioluminescence has recently gained popularity as one of the primary tools for monitoring environmental cleaning.32,35 It has been commonly utilized in the food and pharmaceutical industry; however, limited reports are available for clinical use in healthcare facilities.9 Huang et al. illustrated that ATP bioluminescence is a more sensitive and rapid tool than visual inspection in evaluating the adequacy of cleaning, which is supported by another study that recommended its use to assess the quality of cleaning inside hospitals 9,35. Another study evaluated the adequacy of hospital cleaning procedures using ATP bioluminescence which demonstrated that visual assessment is insufficient to ensure the quality of cleanliness and recommended using ATP bioluminescence to improve the cleaning process in the hospital environment.35 A few limitations were reported, such as the lack of a standardized threshold and the unstudied possibility of interfering chemistries with ATP readings.9
To our knowledge, this is the first study in Egypt evaluating the use of ATP bioluminescence cleaning verification in a tertiary healthcare hospital. The study targeted burn patients who are highly vulnerable to hospital-acquired infections. The study has some limitations, such as the small number of recovered total isolates, which would have necessitated a more extended study period. Nonetheless, this was limited in our study by the allowable period to examine the device.
ATP bioluminescence was found to be a simple and rapid tool for assessing environmental cleaning with a positive impact on the microbial isolation rates from clinical and environmental samples. Quick and objective feedback on surface cleanliness is beneficial for infection control.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Mervat G. El Anany, Professor of Clinical and Chemical Pathology, for her valuable assistance in testing by MALDI-TOF.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All data generated and/or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Richcane A, Tay SC, Pius A, Enoch F, Thomas KG, Poku OSP. Bacteriological profile of burn wound isolates in a burns center of a tertiary hospital. J Acute Dis. 2017;6(4):181-186.
Crossref - Jia H, Li L, Li W, et al. Impact of Healthcare-Associated Infections on Length of Stay: A Study in 68 Hospitals in China. Biomed Res Int. 2019;2019:2590563.
Crossref - Assadian O, Harbarth S, Vos M, Knobloch JK, Asensio A, Widmer AF. Practical recommendations for routine cleaning and disinfection procedures in healthcare institutions: a narrative review. J Hosp Infect. 2021;113:104-114.
Crossref - Flouchi R, Elmniai A, El Far M, Touzani I, El Hachlafi N, Fikri-Benbrahim K. Microbiological Monitoring of the Environment Using the “association Rules” Approach and Disinfection Procedure Evaluation in a Hospital Center in Morocco. J Environ Public Health. 2021;2021:7682042.
Crossref - Carling PC, Huang SS. Improving Healthcare Environmental Cleaning and Disinfection Current and Evolving Issues. Infect Control Hosp Epidemiol. 2013;34(5):507-513.
Crossref - Nascimento EA da S, Poveda V de B, Monteiro J. Evaluation of different monitoring methods of surface cleanliness in operating rooms. Rev Bras Enferm. 2021;74(3):20201263.
Crossref - Leas BF, Sullivan N, Han JH, et al. Environmental Cleaning for the Prevention of Infections, 2015. https://www.ncbi.nlm.nih.gov/books/NBK311016/. Accessed May 21, 2022.
- Pyrek KM. Environmental Cleaning and Monitoring for Infection Prevention. 2013. https://kipdf.com/environmental-cleaning-and-monitoring-for-infection-prevention_5ac706b11723ddab361de6a4.html. Accessed May 21, 2022.
- Huang YS, Chen YC, Chen ML, et al. Comparing visual inspection, aerobic colony counts, and adenosine triphosphate bioluminescence assay for evaluating surface cleanliness at a medical center. Am J Infect Control. 2015;43(8):882-886.
Crossref - Beavis KG, Charnot-Katsikas A. Specimen Collection and Handling for Diagnosis of Infectious Diseases. In: McPherson RA, Pincus MR, eds. Henry’s Clinical Diagnosis and Management by Laboratory Methods. 23rd Ed. Elsevier, Philadelphia, PA. 2017.
- Copeland-Halperin LR, Kaminsky AJ, Bluefeld N, Miraliakbari R. Sample procurement for cultures of infected wounds: A systematic review. J Wound Care. 2016;25(4):s4-s10.
https://doi.org/10.12968/jowc.2016.25.Sup4.S4 - Oumokhtar B, El Ouali Lalami A, Benaicha N, Arhoune B, Bono W. Environmental surfaces in healthcare setting: A great potential risk of pathogens transmission. Biomed Res. 2017;28(6):2398-2401.
- Centers for Disease Control and Prevention (CDC), Infection Control Africa Network (ICAN). Best Practices for Environmental Cleaning in Healthcare Facilities: In Resource-Limited Settings, 2019. http://www.icanetwork.co.za/icanguideline2019/. Accessed May 21, 2022.
- Carroll KC, Pfaller MA, Landry ML, et al., eds. Manual of Clinical Microbiology. 12th Ed. American Society of Microbiology, Washington, D.C. 2019.
Crossref - Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 31st Ed. 2021. www.clsi.org.p:+1.610.688.0100;F:+1.610.688.0700; E:[email protected];W:www.clsi.org. Accessed May 21, 2022.
- Buszewski B, Rogowska A, Pomastowski P, Zloch M, Railean-Plugaru V. Identification of microorganisms by modern analytical techniques. J AOAC Int. 2017;100(6):1607-1623.
Crossref - Gong Y, Peng Y, Luo X, et al. Different Infection Profiles and Antimicrobial Resistance Patterns Between Burn ICU and Common Wards. Front Cell Infect Microbiol. 2021;11:681731.
Crossref - Rahim HR. Isolation and Identification of Some Bacteria Contemn in Burn Wounds in Misan, Iraq. Arch Razi Inst. 2021;76(6):1665-1670.
Crossref - Forson OA, Ayanka E, Olu-Taiwo M, Pappoe-Ashong PJ, Ayeh-Kumi PJ. Bacterial infections in burn wound patients at a tertiary teaching hospital in Accra, Ghana. Ann Burns Fire Disasters. 2017;30(2):116-120.
- Azimi L, Motevallian A, Ebrahimzadeh Namvar A, Asghari B, Lari AR. Nosocomial infections in burned patients in motahari hospital, Tehran, Iran. Dermatol Res Pract. 2011;2011.
Crossref - Otta S, Dash J, Swain B. Aerobic bacteriology of burn wound infections. CHRISMED J Heal Res. 2015;2(4):337.
Crossref - El Hamzaoui N, Barguigua A, Larouz S, Maouloua M. Epidemiology of burn wound bacterial infections at a Meknes hospital, Morocco. New Microbes New Infect. 2020;38:100764.
Crossref - Chen YY, Wu PF, Chen CS, Chen IH, Huang WT, Wang F Der. Trends in microbial profile of burn patients following an event of dust explosion at a tertiary medical center. BMC Infect Dis. 2020;20(1):1-11.
Crossref - Prasad N, Labaze G, Kopacz J, et al. Asymptomatic rectal colonization with carbapenem-resistant Enterobacteriaceae and Clostridium difficile among residents of a long-term care facility in New York City. Am J Infect Control. 2016;44(5):525-532.
Crossref - Kiddee A, Assawatheptawee K, Na-udom A, et al. Risk factors for gastrointestinal colonization and acquisition of carbapenem-resistant gram-negative bacteria among patients in intensive care units in Thailand. Antimicrob Agents Chemother. 2018;62(8):e00341.
Crossref - Tran DM, Larsson M, Olson L, et al. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: Risk factors and burden of disease. J Infect. 2019;79(2):115-122.
Crossref - Sikora A, Zahra F. Nosocomial Infections. In: StatPearls. StatPearls Publishing, Treasure Island, FLorida. 2022.
- Obenza A, Cruz P, Buttner M, Woodard D. Microbial contamination on ambulance surfaces: a systematic literature review. J Hosp Infect. 2022;122:44-59.
Crossref - Ali KM, Al-Jaff BMA. Source and antibiotic susceptibility of gram-negative bacteria causing superficial incisional surgical site infections. Int J Surg Open. 2021;30:100318.
Crossref - Wilson APR, Smyth D, Moore G, et al. The impact of enhanced cleaning within the intensive care unit on contamination of the near-patient environment with hospital pathogens: A randomized crossover study in critical care units in two hospitals. Crit Care Med. 2011;39(4):651-658.
Crossref - Weber DJ, Rutala WA, Kanamori H, Gergen MF, Sickbert-Bennett EE. Carbapenem-Resistant Enterobacteriaceae: Frequency of Hospital Room Contamination and Survival on Various Inoculated Surfaces. Infect Control Hosp Epidemiol. 2015;36(5):590-593.
Crossref - Hassan R, El-Gilany AH, Abd elaal AM, El-Mashad N, Azim DA. An overview of healthcare-associated infections in a tertiary care hospital in Egypt. Infect Prev Pract. 2020;2(3):100059.
Crossref - Mulvey D, Redding P, Robertson C, et al. Finding a benchmark for monitoring hospital cleanliness. J Hosp Infect. 2011;77(1):25-30.
Crossref - Boyce JM, Havill NL, Dumigan DG, Golebiewski M, Balogun O, Rizvani R. Monitoring the Effectiveness of Hospital Cleaning Practices by Use of an Adenosine Triphosphate Bioluminescence Assay. Infect Control Hosp Epidemiol. 2009;30(7):678-684.
Crossref - Zambrano AA, Jones A, Otero P, Ajenjo MC, Labarca JA. Assessment of hospital daily cleaning practices using ATP bioluminescence in a developing country. Braz J Infect Dis. 2014;18(6):675-677.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.