ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.2.33 | © The Author(s). 2019
The current study amid to investigate association of Interleukin-17A with uropathogenic Escherichia coli among patients with urinary tract infection in karbala province, Iraq. Bacterial infections are widespread in urinary tract infections with a global extention. Uropathogenic E.coli (UPEC) is the most common causes of these infections. Out of 110 patients were examined by urologists for urinary tract infection, 25 patients showed positive result for UPEC and other 25 showed positive result for other bacterial pathogens. UPEC were diagnosed depended on the cultural, microscopical, biochemical examinations and confirm the identification by using Vitek2 system. Polymerase chain reaction was used to detection of four genes (pap C, cnfA, fim H, and fyu A). Interleukin-17A concentration in urine was measured by using ELISA kit. out of 110 urine samples, 56 (44.90%) with significant bacteriuria, 44(40%) with non-significant bacteriuria and 10 (9.09 %) with negative culture. The presence of UPEC among significant bacteriuria was 25/56 (44.64 %). The distribution of pap C, cnfA, fim H, and fyu A genes among UPEC were 17(68%), 17(68%), 16(64%) and 15(60%) respectively. Through UTI patients, 50 gave positive (121.70) pg/ml results compared to 30 of control (13. 94) pg/ml. Among uropathogenic Escherichia coli patients, 25 gave positive (92.80) pg/ml results, while 25 of other bacterial pathogens gave positive (15.40) pg/ml results.
Uropathogenic E. coli, Virulence genes, Molecular diagnosis, IL-17A.
Urinary tract infections (UTIs) are the most common type of infection in the human1. Escherichia coli (E. coli) are responsible for about 80% of these infections that have not passed the urinary bladder2. Uropathogenic E. coli has been seen to employ multiple virulence factors that facilitate the colonization and infected the urinary tract. These virulence factors involve adherence molecules (FimH and PapC adhesion molecules)3, iron chelating molecule (like FyuA molecule) that bind ferric iron, and finally secreted toxin like alpha-hemolysin and cytonecrotizing factor 1 (CNF 1)4. One of the most immune defenses against uropathogenic E.coli is the IL-17A. It is upregulated from six hours to one week after inoculation, and remained over the baseline through the two-week of experimental duration5. Interleukin-17A plays role in the innate immune response of infection by enhancing neutrophil migration to infected tissue6. The aim of the study was to evaluate the role of IL-17A as the immunological marker and fimberial adhesin of UPEC in patients with urinary tract infections.
Patients
One hundred and ten patients were clinically diagnosed that have UTIs in the outpatient urology clinic in Imam AL-Hussain Teaching Hospital in Karbala City/Iraq. The study was performed between September 2017 to the February 2018.
Diagnosis of bacteria
All urine samples were cultured on MacConkey agar and incubated overnight at 37°C. E. coli identification was done depending on morphological features biochemical tests (methyl red, indole, Voges-Proskauer, Simmons citrate and urease production)7 and confirmation by using Vitek-2 system (bioMerieux) .
Molecular diagnosis of uropathogenic E.coli genes
Extraction of the DNA
Bacterial DNA was extracted by using commercial kit from Geneaid manifecture’s (Korea). The primers for (pap C, cnfA, fim H, and fyu A) genes were according to8,9,10. The sequence of the primer and amplification condition of PCR as in Table 1.
Table (1):
Primers sequence and PCR amplification conditions.
Genes |
Primer sequence (5′-3′) |
PCR condition |
Size (bp) |
Reference |
|---|---|---|---|---|
papC |
F-GACGGCTGTACTGCAGGGTGTGGCG
R-ATATCCTTTCTGCAGGGATGCAATA |
94°C/5min 1x
94°C/2min 65°C/1min 25x 72°C/2min 72°C/7min 1x |
228 |
Le bouguenec
et al., 1992 |
fimH |
F-TGCAGAACGGATAAGCCGTGG
R-GCAGTCACCTGCCCTCCGGTA |
94°C/5min 1x
94°C/1min 63°C/30s 30x 72°C/3min 72°C/3min 1x |
508 |
Obata-Yasuoka et
al., 2002 |
cnfA |
F- GCAGTCACCTGCCCTCCGGTA
R-CATTCAGAGTCCTGCCCTCATTATT |
95°C/3min 1x
95°C/1min 60°C/1.5min 35x 72°C/3min 72°C/8min 1x |
498 |
Johnson and
Stell, 2000 |
fyuA |
F-TGATTAACCCCGCGACGGGAA
R-CGCAGTAGGCACGATGTTGTA |
95°C/3min 1x
95°C/1min 60°C/1.5min 35x 72°C/3min 72°C/8min 1x |
880 |
Johnson and
Stell, 2000 |
Detection of PCR products
PCR products were separated on a 1.5% gel electrophoresis agarose and visualized by ultraviolet light after addition of ethidium bromide.
IL-17A ELISA Test
Urine level of interleukin 17A was determined by using Human Interleukin 17 ELISA Kit (IL-17A, IL17) from CUSABIO/ USA.
Statistical analysis
Data were analyzed using SPSS statistical package version 21. Chi square (x2), were used to determine the incidence of UPEC among significant bacteriuria. t- test was used to estimate the significant level of the IL-17A28.
Patients and Clinical Isolates
Out of 110 urine samples, 56 (50.90%) found to be with significant bacteriuria and 44(40%) with non-significant bacteriuria and 10 (9.09%) with negative culture. The presence of UPEC among significant bacteriuria in this study was (25/56) 44.64% and 31 (55.36%) other bacterial pathogens as shown in (Fig. 1).
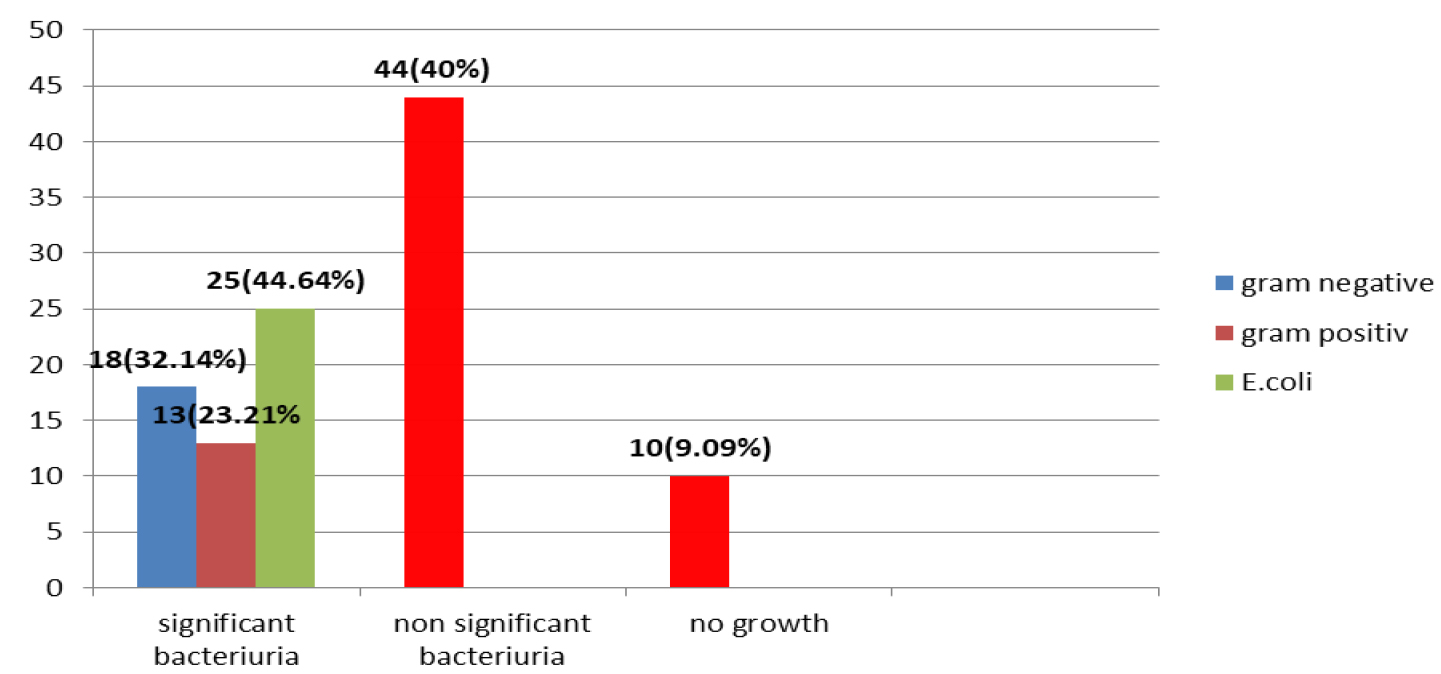
Fig. 1. The percentage of significant bacteriuria, non-significant bacteriuria and negative culture.
UPEC is the main causative agent of community-acquired UTIs which colonize the urinary bladder using a different virulence agents that play important roles in the pathogenesis of UTI4 these involve surface antigenic structures, such as lipopolysaccharide (LPS), polysaccharide capsule, flagella, outer-membrane vesicles, pili, curli, non-pilus adhesins, as well as secreted toxins, secretion systems, and TonB-dependent iron-uptake receptors11.
The results show significant difference (P < 0.05) between the incidence of UPEC and other pathogens among significant bacteriuria. The significant bacteriuria among results was (50.90%) of specimens. These results were higher than results of12 and13, whose found that the significant bacteriuria were (22.7%) and (20.8%) respectively, while, lower than result of14, who reported that the significant bacteruria was (54.80%). As expected E. coli was the most frequently encountered species in the present study (44.64%) which was in the same line with the study conducted in Najaf city by13, who found that E.coli (41.3%) was the predominant causative agent of UTI. On the other hand, these results were lower than the results of15,16,17, whose found that the presence of UPEC were (58.57%), (57.7%) and (67.6%) respectively, while higher than the result of18, who revealed that the presence of E.coli was (33.8%). The differences of these results may be due to different environmental condition and sample size. The highest isolation rate of Escherichia coli infection in the urinary tract may be attributed to the contaminated with fecal flora such as virulence E. coli that facilitate the ascent of bacteria from fecal flora, up the urethra in to the bladder. The urine negative culture may be attributed to other infections such as viral or fungal infections or may be due to recent use of antibiotic19.
Molecular characterization of UPEC virulence factors
There was significant difference between the incidence of UPEC and other pathogens at P< 0.05. Polymerase chain reaction was used in the current study to identify four potential virulence genes (papc, fimH, fyuA ,and cnfA) in uropathogenic E.coli (Fig. 2-5). Out of 25 uropathogenic E.coli, only 5(20%) have all four genes, while the percentage of papc, fimH, cnfA , and fyuA were 17(68%), 16(64%), 17(68%) and 15(60%) respectively (Table 2). These genes were present at (328, 508, 498 and 880) bp respectively in PCR amplification.
Table (2):
The distribution of uropathogenic E.coli virulence genes.
Genes |
Positive |
Negative |
|---|---|---|
papC |
17(68%) |
8(32%) |
fimH |
16(64%) |
9(36%) |
cnfA |
17(68%) |
8(32%) |
fyuA |
15(60%) |
5(40%) |
All genes |
5(20%) |
20(80%) |
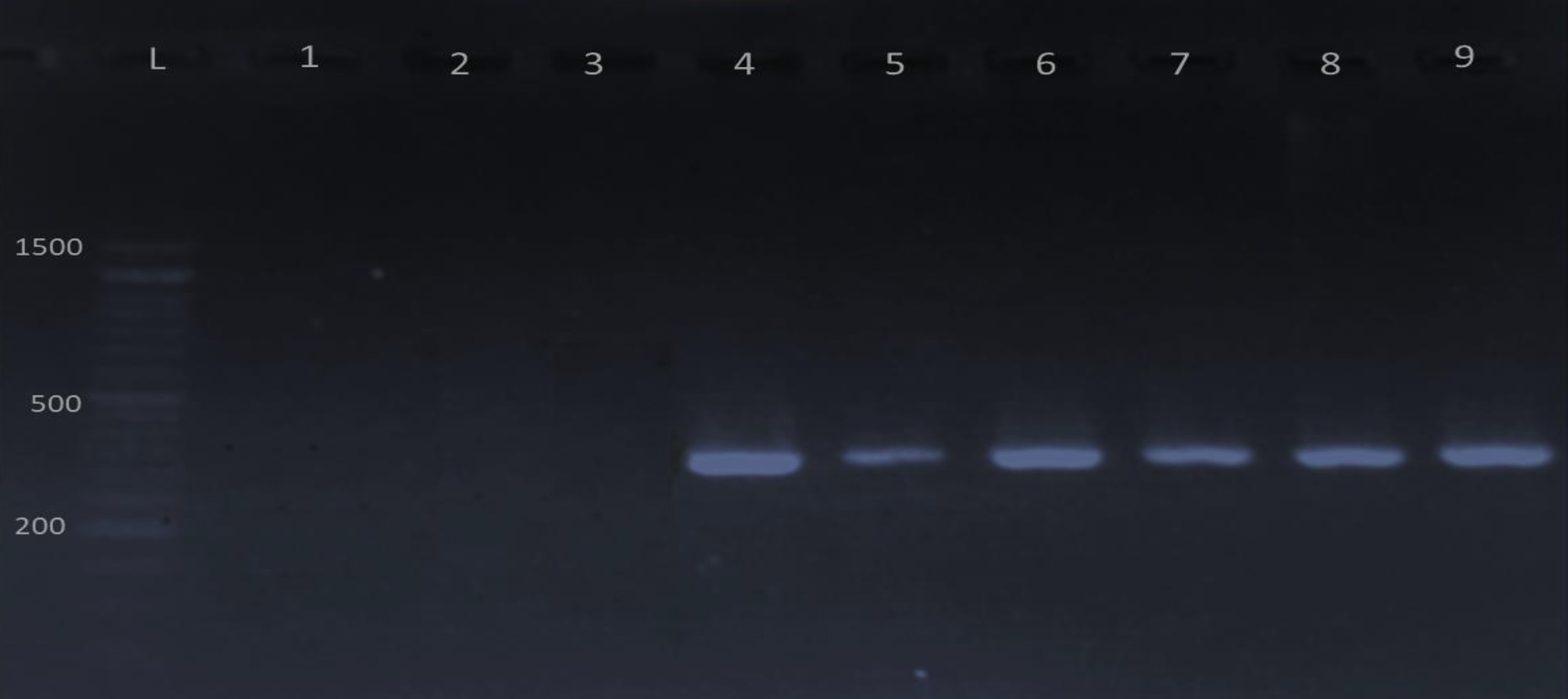
Fig. 2.Gel electrophoresis of PCR products of papC gene visualized under U. V light after mixed with ethidium bromide. L: 1500 bp DNA marker; lane 4-9 positive for papC gene and lane 1-3 negative results. The product size is 328 bp.

Fig. 3.Gel electrophoresis of PCR products of fim H gene visualized under U. V light after mixed with ethidium bromide. L: 1500 bp DNA marker; lane 2-9 positive for fim H gene and lane 1 negative result. The product size is 508 bp.

Fig. 4.Gel electrophoresis of PCR products of fyuA gene visualized under U. V light after mixed with ethidium bromide. L: 1500 bp DNA marker; lane 2-9 positive for fyuA gene and lane 1 negative result. The product size is 880 bp.
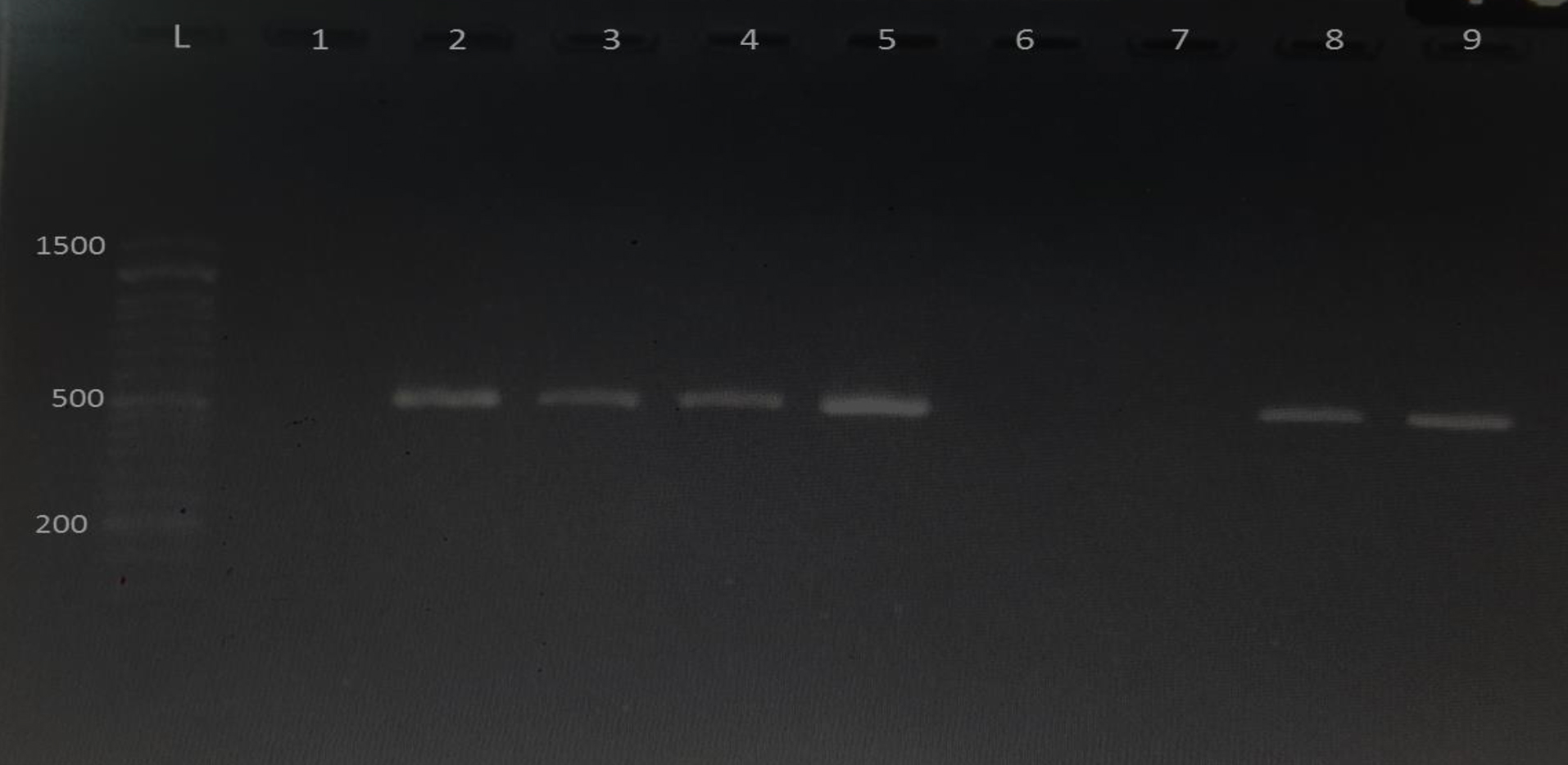
Fig. 5.Gel electrophoresis of PCR products of cnfA gene visualized under U. V light after mixed with ethidium bromide. L: 1500 bp DNA marker; lane 2-5,8,9 positive for cnfA gene and lane 1, 6,7 negative result. The product size is 498 bp.
A genotypic assay was used to detection of different virulence factors of uropathogenic Escherichia coli such as adhesion-encoding genes and other virulence factors that can also associated to virulence in the urinary tract infection. In the present study, these adhesive agents, P fimbriae represents the most common adhesin in UPEC followed by type I fimbriae (fimH)20. The FimH adhesin mediates the bacterial adherence and invasion of host cells and associated with the development of intracellular biofilms formation by UPEC21. The current study were differed from the study of22, who found that fimH (97%) and papC (43%) in UPEC isolates respectively. The second highest prevalence belongs to cytotoxic necrotizing factor A (68%), which encoded by cnf1 gene. The current study result in the same line with study conducted by23, which revealed that the prevalence of cnf gene with the rate (67%). But the present results were lower than the result of24, who found that the prevalence rate for cnf was (79.67%).
In this study, the percent of ferric Yersiniabactin uptake (fyuA) gene was (60%). The results of this study were lower than a study conducted by22, who reported that the percentage of fyuA gene was (83%) in UPEC. Also25, revealed that fyuA gene was (94%) which is also higher than result of current study. The differences in UPEC virulence genes prevalence may be attributed to the climate of different geographic region23,24.
Serum Level of Interleukin -17A
In present study among 50 patients of urinary tract infection compared to 30 of control, The IL-17A mean urine level of patients with UTI were (121.70) pg/ml, but it was (13.94) pg/ml for control and E.coli patients were (92.80) pg/ml, but it was (15.40) pg/ml for other bacterial pathogens patients. There was highly significant difference (p <0.001), (Table 3 and Table 4).
The role of cytokine IL-17A was characterized during urinary tract infection caused by UPEC. The result showed that the men of IL-17A were higher in UTI caused by uropathogenic E.coli than UTI caused by other pathogens and in control groups. This results was agreed with result of studies on mice, and because many of the genes influenced by IL-17A have the same function during UTI in mice and humans26, we expect that all IL-17A results in mice also have the same causes during UTI in humans. IL-17A which is an immune-modulatory cytokines, considers as an innate immune response important factor in the UTI associated with UPEC27. IL-17A contributed to innate clearance of UPEC through a mechanism involving secretion of cytokines and chemokine and cells influx such as neutrophils and macrophages to the bladder5. The gd-positive cells is the key source of IL-17A production27.
In the study of murine model, the mice that lacking of IL-17A display deficient cytokine transcript up regulation and cellular responses during acute urinary tract infection, resulting in suboptimal clearance of uropathogenic E. coli, This clearance defect is likely a result of deficient cytokine and chemokine transcription and impaired of macrophage and neutrophil influx during infection, this result detect IL-17A seems to be unnecessary for the generation of protective immunity, and also IL-17A seems to play a role in regulating the response of innate immunity to UTI27. IL-17A transcript is up regulated in response to acute bladder infection by uropathogenic E. coli time points during a 28-day period, and remains elevated throughout two weeks. A study on infected mice with uropathogenic E. coli– reported increased in the IL-17A, with median values peaking at 48 hours post infection, while other cytokines will be retained to near base line level one week’s post infections, refers to a role for IL-17A in the response of innate immunity to UTI5,27.
Table (3):
IL-17A urine level in Patients with UTI and controls
Subject |
Number |
IL-17A mean
(pg /ml ± SE*) |
|---|---|---|
UTI patients |
50 |
(121.70 ± 22.14)** |
Control |
30 |
(13.94 ±1.77) |
* SE (Standard Error), **Significance (P < 0.001)
Table (4):
IL-17A urine level of E.coli and other pathogens patients.
Subject |
Number |
IL-17A mean
(pg /ml ± SE*) |
|---|---|---|
E.coli patients |
25 |
(92.80±10.05 )** |
Other bacterial pathogens |
25 |
(15.40±4.70 ) |
* SE (Standard Error), **Significance (P < 0.001)
The current study is the first study that revealed the association between human urine level of Interleukin -17A and Escherichia coli in Iraq and the world. The results of current study confirmed that the urine level of Interleukin 17A for E.coli patients were more than that in patients with other bacterial pathogens and control groups.
Acknowledgments
The author is thankful to the Department of Microbiology, College of Medicine, Karbala University, Iraq for the cooperation in the fulfillment of the work. I am also thankful Huda Hadi and Duaa Ali for their cooperation.
Conflicts Of Interest
The authors declare that there are no conflicts of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Schulz L., Hoffman R., Pothof J., Fox B. Top Ten Myths Regarding the Diagnosis and Treatment of Urinary Tract Infections. The Journal of Emergency Medicine, 2 016; 1-6.
- Reisner A., Maierl M., Jצrger M., Krause R., Berger D., Haid A., Tesic D., Zechner E.L. Type 1 fimbriae contribute to catheter-associated urinary tract infections caused by Escherichia coli. J. Bacteriol, 2014; 196(5): 931-9.
- Ron E.Z. Distribution and evolution of virulence factors in septicemic Escherichia coli. Int. J. Med. Microbiol., 2010; 300: 367–370
- Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol., 2015; 13: 269–284.
- Ingersoll M.A., Kline K.A., Nielsen H.V., and Hultgren S.J. G.-C.S.F. induction early in uropathogenic Escherichia coliinfection of the urinary tract modulates host immunity. Cell Microbiol., 2008; 10: 2568-2578
- Iwakura Y., Ishigame H., Saijo S., NakaeS.Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162.
- MacFaddin J.F. Biochemical Tests For Identification of Medical Bacteria, 3rd Edition. Lippincott Williams and Williams, 2000.
- Le Bouguיnec C., Archambaud M., Labigne A. Specific detection of the pap, afa, and sfaadhesin-encoding operons in uropathogenic Escherichia colistrains by polymerase chain reaction. J Clin. Microbiol., 1992; 30: 189–1193.
- Obata-Yasuoka M., Ba-Thein W., Tsukamoto T., Yoshikawa H., Hayashi H. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology, 2002; 148: 2745–2752
- Johnson J.R., Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000; 181: 261–272.
- Terlizzi ME, Gribaudo G and Maffei ME. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Frontiers in Microbiology. 2017; 8: 1-23.
- Kibret M., Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pacific J. Trop. Biomed., 2014; 4(2): 164- 168.
- Al-Hilali S.A.M. Genetic Affinities of Multiple Drug Resistant Uropathogenic Escherichia coli Isolated from Patients with Urinary Tract Infection in Najaf. PhD thesis, University of Kufa, Faculty of Medicine Department of Microbiology, 2015.
- Al-Hasnawy H.H., Jodi M. R. and Hamza H. J. Molecular characterization and sequence analysis of plasmid-mediated quinolone resistance genes in extended-spectrum beta-lactamases producing uropathogenic Escherichia coli in Babylon Province, Iraq. Reviews in Medical Microbiology. 2018, 29:129–135
- Alsamarai A.G.M., Ali S. Urinary tract infections in Kirkuk, Iraq: comparison between pregnant women, diabetic women and female student. WJPPS: in press 2016.
- Al-Kuriashy H.J., Al-Thwani N.A., Khaki I.A. Phenotypic detection of some virulence factors of uropathogenic Escherichia coli isolated from recurrent urinary tract infection patients. Journal of Wassit for Science and Medicine, 2013; 6: 192-200.
- Mohsen T.R., Yano T., Leite D.S. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Rev. Inst. Med. Trop. Sao Paulo, 2015; 50: 255–60.
- Saeed H.C., AL-Otraqchi B.K., Mansoor Y.I. Prevalence of urinary tract infections and antibiotics susceptibility pattern among infants and young children in Erbil city. ZancoJ.Med. Sci., 2015; 19: 915-922.
- Stuthers S., Scanlone J., Parker K., Goddard J., and Hallett R.Parental Reporting of smelly urin and urinary tract infection. Aech Dis Child. 2003, 88(3):250-252.
- Slavchev G., Pisareva E., Markova N. Virulence of Uropathogenic Escherichia coli. J. Culture Collections, 2009; 6: 3-9.
- Wiles T.J., Kulesus R.R., Mulvey M.A. Origins and Virulence Mechanisms of Uropathogenic Escherichia coli. Exp. Mol Pathol, 2008; 85(1): 11-19.
- Cooke N.M., Smith S.G., Kelleher M., Rogers T.R. Major Differences Exist in Frequencies of Virulence Factors and Multidrug Resistance between Community and Nosocomial Escherichia coli Bloodstream Isolates. J Clin. Microbiol., 2010; 48(4): 1099-1104.
- Rasol S.H. Identification of a Number of Escherichia Coli Virulence Genes from Urine Samples in UTI Patients using SSR-PCR Technique, in Duhok and Zakho. MSc. Thesis, College of Medicine, University of Duhok, Iraq, 2013.
- Karimian A., Momtaz H., Mahbobe Madani M. Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr. J. Microbiol. Res., 2012; 6(39): 6811–6816.
- Johnson J.R., Scheutz F., Ulleryd P., Kuskowski M.A., O’Bryan T.T., Sandberg T. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J. Clin. Microbiol, 2005; 43: 3895-900.
- Ragnarsdo´ttir B., Fischer H., Godaly G., Gronberg-Hernandez J., Gustafsson M., Karpman D., Lundstedt A.C., Lutay N., Ramisch S., Svensson M. L., et al. TLR- and CXCR1-dependent innate immunity: insights into the genetics of urinary tract infections. Eur. J. Clin. Invest., 2008; 38(2): 12–20.
- Sivick K.E., Schaller M.A., Smith S.N, and Mobley H.L.T. The Innate Immune Response to Uropathogenic Escherichia coli Involves IL-17A in a Murine Model of Urinary Tract infection. J. Immunol, 2010; 184: 2065-2075.
- Walker G.A. and Shostak J. Common statistical methods for clinical research with SAS Examples, 3rd edn, SAS. Institute Inc., Cary, North Calorina, USA, 2010; 539.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


