ISSN: 0973-7510
E-ISSN: 2581-690X
The subsequent application of insecticides and pesticides in agriculture results in several health and environment issues. To overcome, such devastating effect of synthetic chemicals, an eco friendly measure is required. Chitinolytic bacteria and their enzymes can be adopted as potent substitutes to chemicals required controlling the agriculture loss. The aim of this research was to isolate bacterial strains with significant chitinase activity. Isolates were screened based on method viz. zone inhibition, colorimetric, biochemical and were identified based on 16S rDNA sequences. Observed chitinase activity was in range 0.181Uml-1 to 1.594Uml-1 with zone inhibition in the range of 6mm to 29mm. Among the recovered strains only two, MSCP10 and MSCW8 showed good response when tested against insect and showed 80% and 95% mortality respectively after 72 hours of treatment. Based on 16S rDNA sequencing, MSCP10 and MSCW8 exhibited similarity with Serratia marcescens strain S308 and Staphylococcaceae bacterium HDMd_5 respectively. Through insect bioassay it was concluded that these bacterial strains were effective against Lepidopteran insect P. xylostella.
16S rDNA sequencing, Chitinolytic bacteria, Chitinase, Plutella xylostella.
The use of naturally occurring chitinolytic bacteria, actinomycetes and fungi as potential supplements for insecticides pesticides have been reported in many studies1. Chitin, the second most abundantly found and widely distributed natural renewable resource next to cellulose in nature. It is a homopolymer of ß-1, 4-linked N-acetyl-D-glucosamine. Chitin is the main structural component of shells of crustaceans, exoskeleton of insects, fungal cell wall and protozoa. The annual worldwide turnover of chitin is around 100 billion tons2. Based on amino acid sequences present chitinases are the enzymes that catalyze chitin degradation and divided into Family 18 and 19 of glycosyl hydrolase1. Several bacteria produces chitinases to degrade chitin and utilize it as an energy source and thereby helpful in recycling these resources in soil ecosystem2.
A large number of chitinolytic soil bacteria have been isolated from soil3, shellfish waste4, shrimp shell-enriched soil5 and vermicompost6. Phytospheres, such as rhizosphere and phylloplane, are important habitats for chitinolytic bacteria7,8 . There is a considerable interest in chitinolytic bacteria for efficient bioconversion of chitinaceous waste based on the exploitation of chitinases. Soil bacteria are excellent sources of chitinases and could be used for catabolic conversion of chitinaceous waste into useful molecules for application in agriculture, biotechnology and medicine9,10. Bacteria from genera like Bacillus, Serratia, Pseudomonas, Streptomyces and Aeromonas frequently occur in soil and are potentially suitable sources of enzymes. Recycling of chitinous waste using chemical treatments is a costly process11. The continous use of chemicals in agriculture leads to numerous environment and public health problems. Hence, there is a need to look forward for an environmentally sound and cost effective approach. The use of chitinolytic bacteria and chitinases (exochitinase and endochitinase) can be adopted as an alternative to both, degradation of chitinous waste and as biocontrol agent.
In the present study, bulk soil, rich in chitinous waste and rhizospheric soil rich in microbes’ were collected randomly for isolation, screening of chitinolytic bacteria and performed insect mortality bioassay with selected strains.
Local market (fish and chicken) of Delhi NCR and rhizospheric regions of Ludhiana and Meerut were randomly selected for soil collection (Table 1). Soil sampling was done using the quadrat method of sampling and was processed.
Table (1):
Location of collected soil sample.
Type of sample |
Location of sample |
|---|---|
Rhizospheric soil |
Sugarcane crop, Ludhiana |
Rhizospheric soil |
Wheat crop, Ludhiana |
Non-rhizospheric soil |
Industrial area of Meerut |
Non-rhizospheric soil |
Fish and poultry market Delhi-NCR |
Preparation of colloidal chitin
Colloidal chitin was prepared by method described by Roberts and Seltrennikoff with modification12. To 2gm of chitin powder 35ml of concentrated HCl was added, incubated overnight at 4°C and next day ice cold 200ml ethanol was added to the mixture, stand overnight at room temperature and centrifuge it at 10,000(g) for 30min. The mixture was filtered through fine muslin cloth with continuous washings of distilled water. Recovered colloidal chitin was stored at 4°C until use.
Isolation of chitinase producers
Chitinolytic bacteria from collected soil samples were isolated by serial dilution and spread plate method. On each plate 0.1ml of dilution was plated in triplicates on minimal salt media (MSM) containing Na2HPO4.2H2O (3.5g), KH2PO4 (1.0g), (NH4)2SO4 (0.5g), MgCl2.6H2O (0.1g), Ca (NO3)2.4H2O (0.05g) and chitin (5.0g) as carbon source and incubated at 30°C for 3days. The total 28 chitinase producers were selected based on the morphology, color and area of clearance around the colonies.
Screening of chitinase producing bacteria
Quadrant streak of all the isolates were carried out on MSM plate amended with 0.5% chitin to isolate the potential bacteria based on the chitinase produced.
Further, isolates were screened using different concentrations of chitin (0.5, 1.0, 1.5, 2.0 and 2.5%) in MSM plates and incubated at 30°C for three to five days. Colonies with larger clear zone size (>=10mm) were selected. The pure isolates were preserved in chitin containing nutrient broth glycerol stock at -80°C to maintain viability.
Characterization of Bacterial isolates
Identification of chitinolytic bacterium
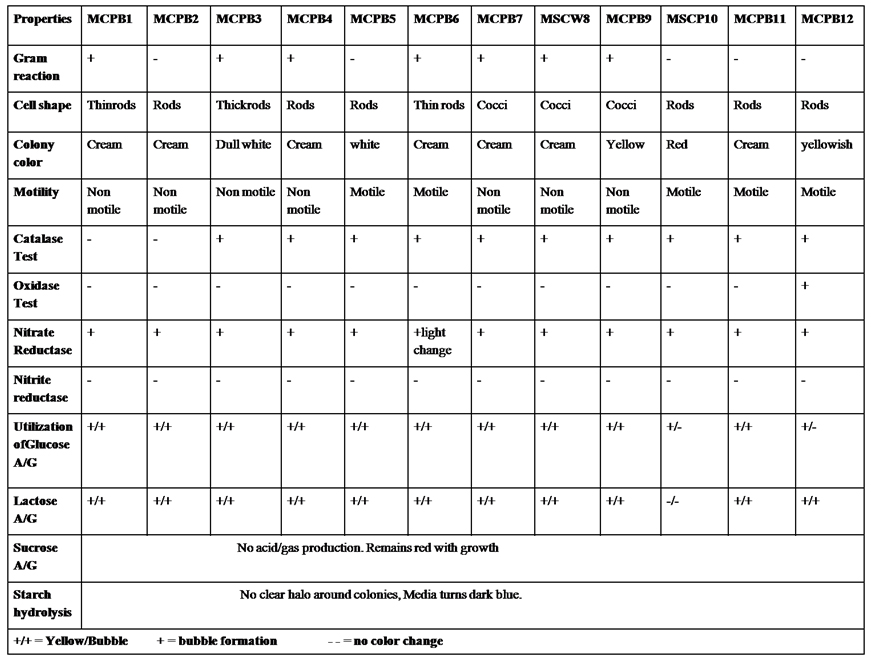
The isolates were identified through their morphological and physiological properties according to Bergey’s manual of systematic bacteriology13 (Table 2).
Chitinase assay
Colorimetric method described by Setia and Suharjona was used to determine the chitinase activity with three replications14. The reaction mixture consists of 1ml crude enzyme, 1.5ml of 1% colloidal chitin substrate in 200mM (pH6) potassium phosphate buffer. The reaction mix was incubated at 30°C for 2 h and boiled for 10 min to stop the reaction. Then centrifuge at 8000rpm for 20 min. collected the supernatant and added 1ml of Dinitrosalicylic acid (DNS) in 1ml of supernatant boiled for 5 min and left at room temperature to cool down. Absorbance was measured at 540nm against the standard curve of N-acetylglucosamine (GlcNAc) plotted between GlcNAc concentrations and GlcNAc absorbance values. One unit of chitinase activity was defined as the amount of enzyme required to liberate 1.0mg of GlcNAc per h.
PCR amplification of genomic DNA for 16S rDNA sequencing
The total genomic DNA was isolated from the samples. Approximately (1µl DNA) 1.3/1.5Kb, 16S rDNA fragment was amplified using high fidelity PCR polymerase. The PCR products were sequenced bi-directionally using 16S forward and reverse primer 5’-AGHGTBTGHTCMTGNCTCAS-3’and 5’-TRCGGYTMCCTTGTWHCGACTH-3’ respectively using gradient polymerase chain reaction (ABI 3500 Genetic Analyzer). The PCR amplification was performed with initial denaturation (96°C; 5min), denaturation (96°C; 30s), hybridization (50°C; 30s) and elongation (60°C; 1.30min). The PCR amplified product was analysed on 1% agarose gel and with 500bp ladder. The amplified sequence was analysed using Data analysis software (seq Scape- v 5.2).
Phylogeny tree construction
For the construction of phylogeny, 16S rDNA sequences was matched with reference strains sequences (Table 3) in Genebank database (http://www.ncbi.nlm.nih.gov) and was aligned using Clustal W Multiple Alignment tool in MEGAV.7 program.
Table (3):
Reference strains for construction of phylogeny based on 16S rDNA sequences.
| Insect: Plutella xylostella (10d old) No. of Larvae: 10 |
||
|---|---|---|
| 1% Bacterial stock (~cell count 1.5X10^8 CFU/ml) | Treating concentration | Mortality rate after 3days of treatment (in percent) |
| MCPB1 | 50µl | 0 |
| MCPB2 | 50µl | 5 |
| MCPB3 | 50µl | 10 |
| MCPB4 | 50µl | 25 |
| MCPB5 | 50µl | 10 |
| MCPB6 | 50µl | 15 |
| MCPB7 | 50µl | 85 |
| MSCW8 | 50µl | 95 |
| MCPB9 | 50µl | 70 |
| MSCP10 | 50µl | 80 |
| MCPB11 | 50µl | 75 |
| MCPB12 | 50µl | 40 |
| Control (Distilled Water) | 50µl | 0 |
Statistical analysis
The standard statistical software Graph Pad Prism was used to carry out the data analysis. The mean and standard deviation were used to summarize the collection of data for each measurement. Two-way analysis of variance was used to evaluate the influence of independent bacterial strains. Bonferronimultiple comparision procedure was used to determine whether the data show evidence of difference between the various classes of chitinolytic bacteria.
A total of 28 chitinase producing bacterial strains were isolated from the soil samples collected from different sites (Fig. 1). Out of 28 strains only 12 showed clear zone (>=10mm) when incubated in different concentrations (0.5, 1.0, 1.5, 2.0 and 2.5%) containing chitin media plates were selected and labeled as (MCPB1, MCPB2, MCPB3, MCPB4, MCPB5, MCPB6, MCPB7, MSCW8, MCPB9, MSCP10, MCPB11 and MCPB12) (Fig. 2). The formation of clear zone around the colonies indicates the presence of chitinase activity, to utilize chitin as a source of carbon and nitrogen. These finally selected 12 pure isolates were subjected to identification through biochemical tests. Observed results of biochemical tests presented in tabulated form (Table 2) and also compared with each other (i) comparing zone diameter (in mm) using different concentrations of chitin amended MSM plates (Fig. 3) (ii) and measuring chitinase activity (Uml-1) in chitin containing MSM broth (Fig. 4). On the basis of preliminary bioassay data two isolates (MSCW8 and MSCP10) were found to be more potent among 12 isolates and further selected for molecular identification (16S rDNA sequencing). The biochemical characteristics inferred that MCPB1 and MCPB2 shared similar characteristics with Lactobacillus. MCPB3, MCPB4 and MCPB6 with Bacillus. MCPB7, MCPB9 and MSCW8 matches with Staphylococcus and Micrococcus. MCPB5 and MCPB11 were observed similar to Escherichia. MSCP10 and MCPB12 were found similar to Serratia and Pseudomonas respectively.
Fig. 1. Isolation of chitinolytic bacterial strains from soil
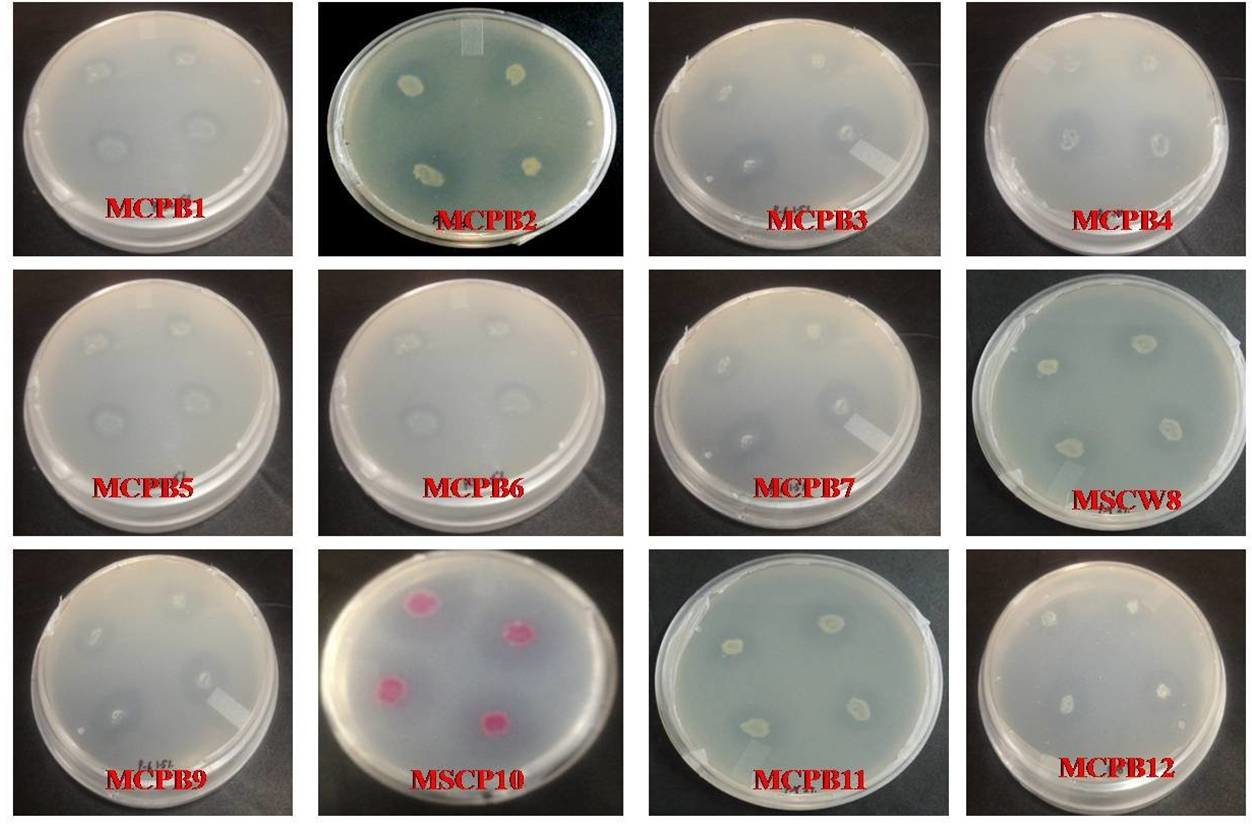
Fig. 2. Selection of different bacterial strains on the basis of zone size (>=10mm) after 72 h of incubation
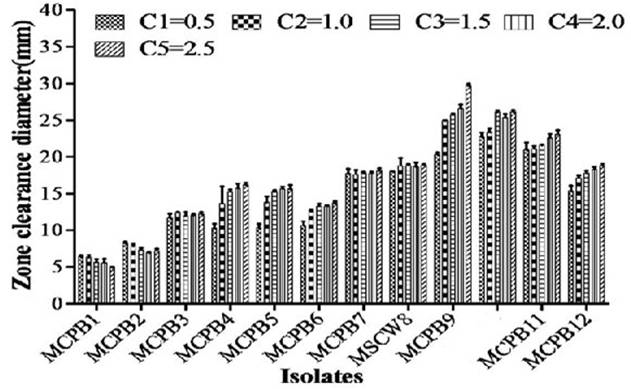
Fig. 3. Zone of inhibition at different concentration of chitin (C1=0.5, C2=1.0, C3=1.5, C4=2.0 and C5=2.5% respectively). Experiment was performed in triplicates, the error bars represents mean ± standard deviation at P value 0.0001<0.01<0.05
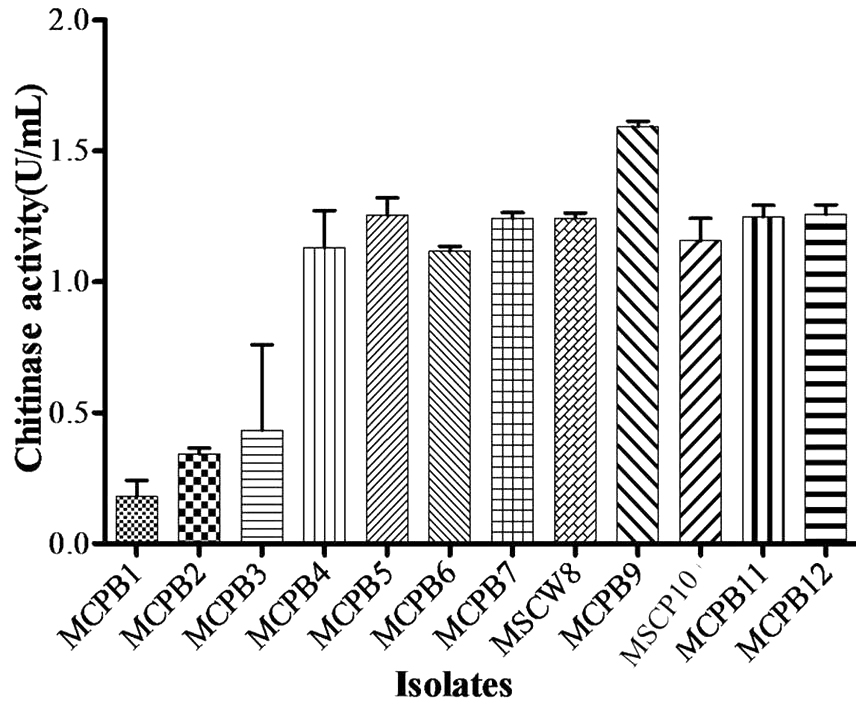
Fig. 4. Chitinase activity of selected bacterial strains. One unit of chitinase activity was defined as the amount of enzyme required to liberate 1.0mg of GlcNAc per h. Experiment was performed in triplicates, the error bars represents mean ± standard deviation at P value 0.001<0.01<0.05
Table (2):
Morphological and Biochemical characterization of selected bacterial strains.
 The amplicon 16S rDNA sequences of isolate MSCW8 and MSCP10 on 1% agarose gel was compared with 500bp ladder (Fig. 5). Amplified 16S rRNA sequences showed that isolate MSCW8 (Accession no. MG066581) and MSCP10 (Accession no. MG066582) were found to be most similar to Staphylococcaceae bacterium HDMd_5 and Serratiamarcescens strain S308 respectively with 99% phylogenetic similarity (Fig. 6, a and b)
The amplicon 16S rDNA sequences of isolate MSCW8 and MSCP10 on 1% agarose gel was compared with 500bp ladder (Fig. 5). Amplified 16S rRNA sequences showed that isolate MSCW8 (Accession no. MG066581) and MSCP10 (Accession no. MG066582) were found to be most similar to Staphylococcaceae bacterium HDMd_5 and Serratiamarcescens strain S308 respectively with 99% phylogenetic similarity (Fig. 6, a and b)
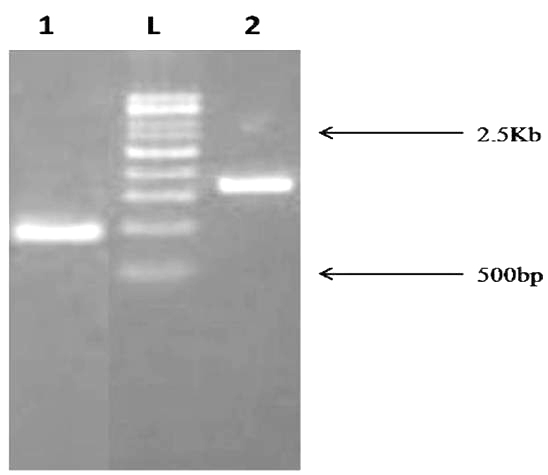
Fig. 5. PCR amplification of 16S rDNA fragment from Bacterial sample. The size of PCR amplified product is ~1.5Kb Lane description: L – 500bp ladder 1. MSCP10 2. MSCW8
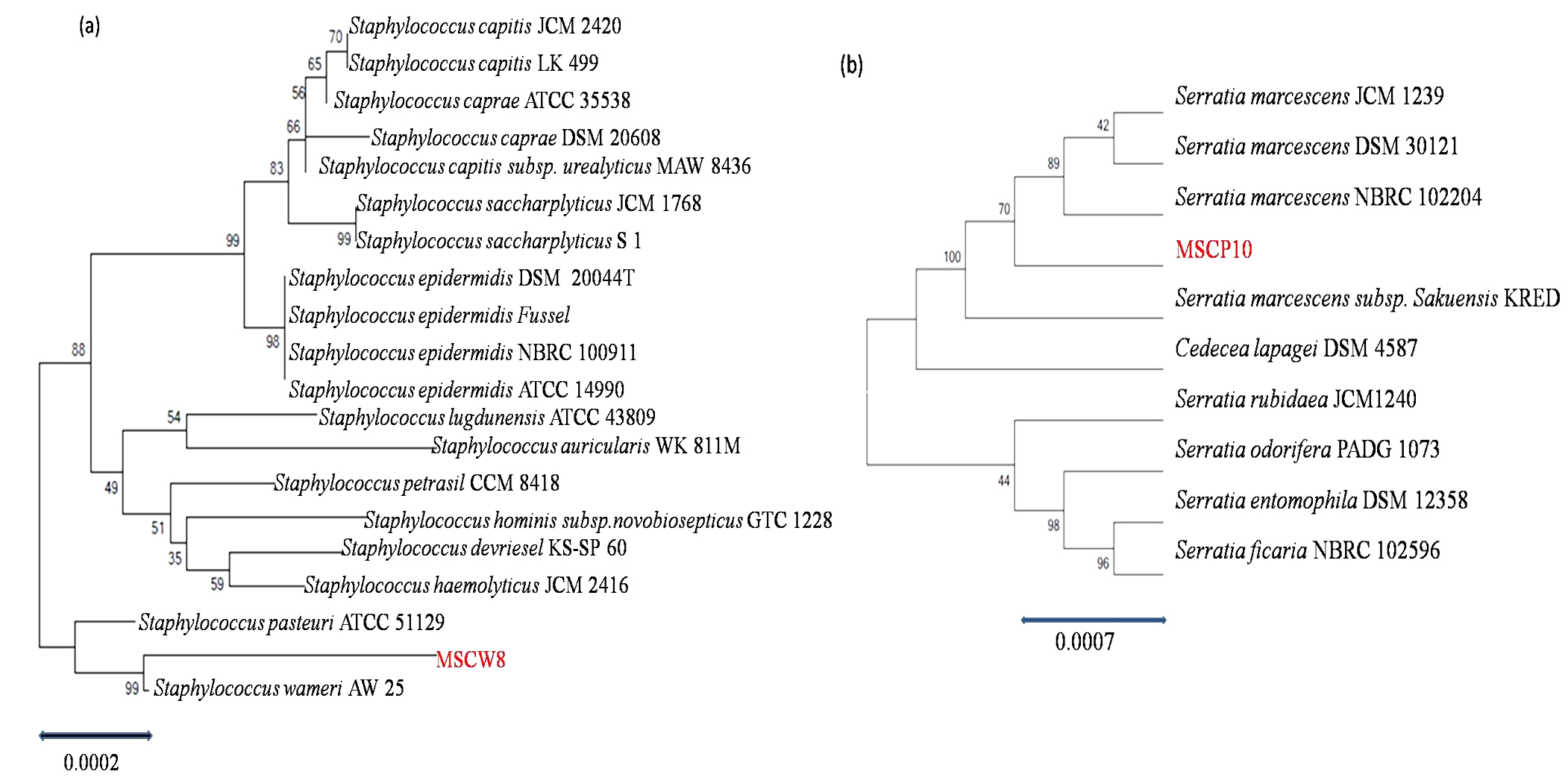
Fig. 6. Phylogeny tree of isolate (a) MSCW8; (b) MSCP10 compared with Genebank database (http://www.ncbi.nlm.nih.gov) and was aligned using Clustal W Multiple Alignment tool in MEGAV.7 program
Total 28 isolates were recovered from chitinous rich bulk soil and agriculture land. These isolates were further screened on the basis of potential to utilize chitin. Out of 12, two efficient isolates (MSCW8 and MSCP10) were identified using 16S rDNA sequence. These isolates were found similar to Staphylococcaceae bacterium HDMd_5 andSerratiamarcescens strain S308 respectively. In present study observed chitinase activity was in range 0.181Uml-1 to 1.594Uml-1. The zone clearance diameter around colonies is in range 6mm to 29mm. This infers that these isolates produce exochitinases for the utilization of chitin. Suharjona and Satia reported highest chitinase activity at 30°C (pH7) after 4 days of incubation in Streptomyces sp. S242 (0.162Uml-1), Bacillus thuringiensis (0.23Uml-1), Serratia marcescens DSM3012 (0.556Uml-1)14. Other chitinase producing different species of Bacillus were previously mentioned such as B. amyloliquefaciens15, B. cereus16, B. licheniformis17, B. megaterium18, B. circulans19, B. subtilis20, B. thuringiensissub sp.aizawai21, B.stereothermophilus22 and B. thuringiensissub sp. krustaki23. Present research was employed with the aim of utilizing chitinase producers as an efficient biocontrol agent. Mubarik et al. reported chitinase from Bacillus sp. as biocontrol agent24. Isolate MSCP10 and MSCW8 shows effectiveness against Plutella xylostella larvae in preliminary bioassay as observed (Table 4). After 72 h of treatment larval mortality can be easily observed. Different concentrations of isolates were mixed with the diet and larvae were allowed to feed upon. As bacteria starts multiplying in the gut of larvae, it puncture the gut lining, larvae feels pain in stomach and feed less. As a result chitinase start degrading the gut lining of larvae made of chitin. From above conducted experiment we can interpret that chitinolytic bacteria as such can be used as biocontrol agent inspite of using simple spray of chitinase directly in field or in combination with delta toxin was found to be more effective25,26 which is a long and tedious process. Application of chitin to a plant was an effective biocontrol agent for pest insects, as it attracts chitinolytic bacteria to produce chitinase27.Table (4):
Bacterial strains bioassay with P.xylostella larvae.
Chitin degrading bacteria are promising and versatile agent in the field of agriculture, industry, medicine and other commercial uses. From current study data it can be concluded that isolated chitinolytic bacterial strains were effective against Lepidopteran insect P. xylostella. Further confirmation can be achieved by conducting insect bioassay with recovered strains in controlled laboratory conditions and consecutive field trials.
ACKNOWLEDGMENTS
The author would like to thank Division of Entomology, ICAR-Indian Agriculture Research Institute, New Delhi for providing insect culture.
- Kamil, Z., Saleh, M., Moustafa, S. Isolation and identification of rhizosphere soil chitinolytic bacteria and their potential in antifungal biocontrol. Global. J. Mol. Sci., 2007; 2: 57-66
- Das, S.N., Sarma, P.V.S.R.N., Neeraja, C., Malati, N., Podile, A.R. Members of Gammaproteobacteria and Bacilli represent the culturable diversity of chitinolytic bacteria in chitin-enriched soils. World. J. Microbiol. Biotechnol., 2010; 26: 1875-1881
- Wang, S.L., Chang, W.T. Purification and characterization of two bifunctional chitinase/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl. Environ. Microbiol., 1997; 63: 380-386
- Wang, S.L., Hwang, J.R. Microbial reclamation of shellfish wastes for the production of chitinases. Enzyme. Microb. Technol., 2001; 28: 376-382
- Zhu, X.F., Zhou, Y., Feng, J.L. Analysis of both chitinase and chitosanase produced by Sphingomonas sp. CJ-5. J. Zhejiang. Univ. Sci. B., 2007; 8: 831-838
- Yasir, M., Aslam, Z., Kim, S.W., Lee, S.W., Jeon, C.O., Chung, Y.R. Bacterial community composition and chitinase gene diversity of vermicompost with antifungal activity. Bioresour. Technol., 2009; 100: 4396-4403
- Gonzalez-Franco, A.C., Deobald, L.A., Spivak, A., Crawford, D.L. Actinobacterial chitinase-like enzymes: profiles of rhizosphere versus non-rhizosphere isolates. Can. J. Microbiol., 2003; 49: 683-698
- Kishore, G.K., Pande, S., Podile, A.R. Biological control of late leaf spot of peanut (Arachis hypogaea) with chitinolytic bacteria. Phytopathology, 2005a; 95: 1157-1165
- Kishore, G.K., Pande, S., Podile, A.R. Phylloplane bacteria increase seedling emergence, growth and yield of field grown groundnut (Arachis hypogaea L.). Lett. Appl. Microbiol., 2005b; 40: 260-268
- Bhattacharya, D., Nagpure, A., Gupta, R.K. Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol., 2007; 27: 21-28
- Krithika, S., Chellaram, C. Isolation, screening, and characterization of chitinase producing bacteria from marine wastes. Int. J. Pharm. Pharmac. Sci., 2016; 8: 34-36
- Roberts, W.K., Seltrennikoff, C.P. Plant and bacterial chitinases differ in antifungal activity. Microbiology, 1988; 134: 169-176
- Keddie, R.M., Shaw, S., Kurthia, G. In: Bergey’s manual of systematic bacteriology. Sneath PH, Mair N, Sharpe ME, Holt JG. Eds. Baltimore: Williams and Wilkins, 1986; 2: 1255-8
- Setia, I.N., Suharjono. Chitinolytic assay and identification of bacteria isolated from shrimp waste based on 16S rDNA sequences. Adv. Microbiol., 2015; 5: 541-548
- Wang, S.Y., Moyne, A.L., Thottappilly, G., Wu, S.J., Locy, R.D., Singh, N.K. Purification and characterization of a Bacillus cereus exochitinase. Enzyme. Microb. Technol., 2001; 28: 492-498
- Huang, C.J., Wang, T.K., Chung, S.C., Chen, C.Y. Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus 28-9. BMB Reports, 2005; 38: 82-88
- Waldeck, J., Daum, G., Bisping, B., Meinhardt, F. Isolation and molecular characterization of chitinase deficient Bacillus licheniformis strains capable of deproteinization of shrimp shell waste to obtain highly viscous chitin. Appl. Environ. Microbiol., 2006; 72: 7879-7885
- Brzezinska, M.S., Jankiewicz, U., Burkowska, A., Walczak, M. Chitinolytic microorganisms and their possible application in environmental protection. Curr. Microbiol., 2014; 68: 71-81.
- Huang, C.J., Chen, C.Y. High-level expression and characterization of two chitinases, ChiCH and ChiCW, of Bacillus cereus 28-9 in Escherichia coli. Biochem. Biophy. Res. Comm., 2005; 327: 8-17
- Wang, S.L., Lin, T.Y., Yen, Y.H., Liao, H.F., Chen, Y.J. Bioconversion of shellfish chitin wastes for the production of W-118 chitinase. Carbohydr. Res., 2006; 341: 2507-15
- Morales, de, la, Vega, L., Barboza-Corona, J.E., Aguilar-Uscanga, M.G., Ramírez-Lepe, M. Purification and characterization of an exochitinase from Bacillus thuringiensis subsp. aizawai and its action against phytopathogenic fungi. Can. J. Microbiol., 2006; 52: 651-657.
- Sakai, K., Yokota, A., Kurokawa, H., Wakayama, M., Moriguchi, M. Purification and characterization of three thermostable endochitinases of a noble Bacillus strain, MH-1, isolated from chitin-containing compost. Appl. Environ. Microbiol., 1998; 64: 3397-3402
- Driss, F., Kallassy Awad, M., Zouari, N., Jaoua, S. Molecular characterization of a novel chitinase from Bacillus thuringiensis subsp. kurstaki. J. Appl. Microbiol., 2005; 99: 945-953
- Mubarik, N.R., Mahagiani, I., Anindyaputri, A., Santoso, S., Rusmana, I. Chitinolytic bacteria isolated from chili rhizosphere: chitinase characterization and its application as biocontrol for whitefly (Bemisia tabaci Genn.). Am. J. Agr. Biol. Sci., 2010; 5: 430-5
- Koga, D. Application of chitinase in agriculture. J. Met. Mater. Miner., 2005; 15: 33-6
- Patil, R.S., Ghormade, V., Deshpande, M.V. Chitinolytic enzymes: an exploration. Enzyme Microb. Technol., 2000; 26: 473-483
- Metcalfe, A.C., Krsek, M., Gooday, G.W., Prosser, J.I., Wellington, E.M.H. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol., 2002; 68: 5042-5050.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


