ISSN: 0973-7510
E-ISSN: 2581-690X
This research was conducted to identify the bacterial hydrocarbonclastic in waste tanks, Petapahan, Riau, Indonesia. Bacteria hydrocarbonoclastic are bacteria that have traffic in degrade petroleum. Analysis of 16S rRNA used the primers pair 63 Forward and Reverse 1387 produced 1300 bp. Results of the analysis of the 16S rRNA sequence in bacterial isolates obtained that IMB- 09 09 was similar to Pseudomonas tollasii, IMB- 10 was similar to Bacillus cereusIMB 11 had similarities with Bacillus toyonensis strain BCT-7112, IMB 12 was Lysinobacillus fusiform strain NBRC 15717, IMB 15 was Pseudomonas stutzeri strain ATCC 17588 entirely did have the ability to degrade petroleum.
16S rRNA, Hydrocarbonoclastic, Bacteria, Waste tank, Petroleum
As a source of energy, oil and gas have many benefits, it is quite efficient and economical as well as it exists is quite abundant, but when it is spilled or lost to the environment, the oil will be impurities that can become harmful pollutants. Petroleum includes hazardous materials and toxic waste. Petroleum pollution can come from drops and spills of petroleum during the activities of drilling, production, refining, and transportation of oil, resulting in the disruption in the balance of ecosystems, land, water or sea1. One of the contaminants that are difficult to render is the hydrocarbon compounds derived from petroleum or petroleum sludge. These compounds can be toxic if accumulated in the cell.
Biodegradation of hydrocarbons such as petroleum compounds usually require the cooperation of more than one species of bacteria, it is because petroleum is formed from many different hydrocarbon compounds and bacteria can only use hydrocarbon in a specified range. The difference in the ability of the bacteria in the use of hydrocarbon compounds can be used to maximize the biodegradation process, therefore, the ability of a characterization of bacteria can degrade the hydrocarbon compounds became very important to do2,3.
The process of degradated petroleum start from inoculated the bacterias and take interaction with oil. Processing degradation was occure in aerobic conditions. Degradation petroleum in aerobic more quick than in aerobic conditions, because bacterias produce more energy compare anaerobe reactions4. Interaction between microbe and hydrocabron by adhetion or emulsification for eliminate anionic heteropolysacarade at wall cell or capsule. All of precess ware activated by oxigenase enzyme.
Petroleum degradation be occured at peripheral to transform hydrocarbon become intermediet compouns such as asetile CoA. CoA was going to come to threecarbocsilate cycle and produce Co2 + H2O and energy for growth5,6. Many factors take impect in petrileum degradation such as petroleum chemistry compouns, microbials community, themperature, oxigen, pH and nutrients.
Genomic DNA Isolation Using Kit invitrogen
All bacteria samples had the ability to degrade petroleum waste, will be tested and analysis of the 16S rRNA(Figure 1). this method had to mixture all samples in the best liquid media. Nutrient Broth (NB) during the 16 hours at 28 ° C with 120 rpm. Genomic extraction using DNA purification KIT Wizard (Invitrogen) follows the KIT instruction: the first stage of Digestion: 1 ml bacterial suspension 1.5 ml microtube put into the centrifuge bacterial suspension with a speed of 5000 rpm for 5 minutes. Add lysozyme digestion buffer as much as 180 µl vortex, incubation temperature 37oC for 30 minutes. Next add 20 µl proteinase K, vortex and then added 200 µl of Lysis buffer and then bindings genomic vortex and incubated for 30 minutes at a temperature of 55oC, absolute ethanol added as many as 200 µl and vortex.
The second stage of binding: put spin Klum into the tube, pour the liquid from the binding process into spin Klum and centrifuge with a speed of 10.000 rpm for 2 minutes, remove the collection tube, replace with a new one. The third stage of washing: Add 500 µl wash buffer I, centrifuge with a speed of 10.000 rpm for 2 minutes, remove the collection tube, and replaced with a new one. Add 500 µl wash buffer 2, centrifuges with speed 12,000 rpm for 3 minutes, then taken spin Klum, enter into a sterile 1.5 ml micro tube. The fourth stage Elusi: add 200 µl genomic elusion buffer, leave for 1 minute centrifuge with speed 12.000 rpm for 3 minutes, remove spin Klum, next electrophoresis agarose gel with 1.5% and see in UV light.
DNA amplification
Amplificationu sing16sr RNA Primer from Promega, stages that are conducted in accordance with the instructions of the manufacturer of the kit Protocol. DNA had extracted (Eppendorf, Westbury, NY) for 15 µl of 100 minutes then added DNA rehydration (Promega). Amplification of the 16S gene rRNA had done in 50µl containing 10 pmol 63f primer (5 ‘-AGAGTTTGATC (A/C) TGGCTCAG-3 ‘) and 1387r (5 ‘-GG (C/T) TACCTTGTTACGACTT-3 ‘). Around of 25 µl green tag Promega, DNA template 3 µl (10 ng/ µl), the addition of a nuclease-free water 20 µl of PCR Amplification cycle was 35. beginning with the denaturation temperature was 94°C for 3, then proceed with stage 94°C denaturation for 1 minute. 55°C temperature Annealing lasted for 30 seconds, the expansions for 30 seconds at a temperature of 72°C and end satthe end of the expansions at72°C for 5 seconds. PCR cycle last sasmuchas 35cycles, 3 hours. 7 µl of PCR results are examined by gel electrophoresis. Visualization of results of PCR was performed on agarose electrophoresis 75 grin 50 ml of 0,5TBE.
Genomic DNA Isolation Using KIT INVITROGEN
The genomic DNA isolation using DNA isolation kit from INVITROGEN beginning from the taking of the pellet suspension of bacteria in culture liquid (Nutrient broth) using the centrifuge. Isolation of DNA results can be seen from Figure 1 below.

Fig. 1. Genomic DNA isolation
Each sample of DNA isolation result showed the results good enough, this is because the isolation technique that is used is the isolation techniques already adopted from the isolation kit used, namely INVITROGEN. Samples of IMB and IMB-10 shows the results of isolation are not good, look at the picture that the very thin ribbon, possibly this is due to a number of pellets obtained from bacterial suspensions IMB and IMB-10 bit. The results of this DNA isolation continued to amplify with a PCR.
Amplified genomic DNA with primer 16s rRNA
Amplication Genomic DNA with the primer 16s rRNA Amplification using PCR machine Thermo. The PCR cycle run is 35 cycles. The results of PCR with primer 63F and 1387R can be seen in Figure 2 below.
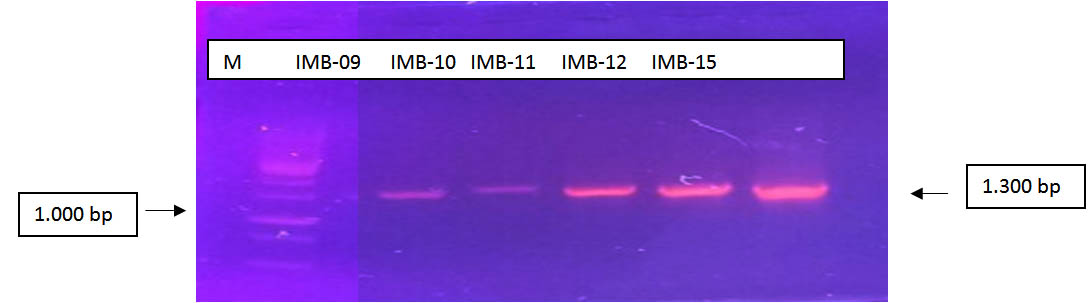
Fig. 2. Results of PCR using primer pairs 63F and 1387R (M = Marker 1000bp; IMB (Isolates Petroleum)
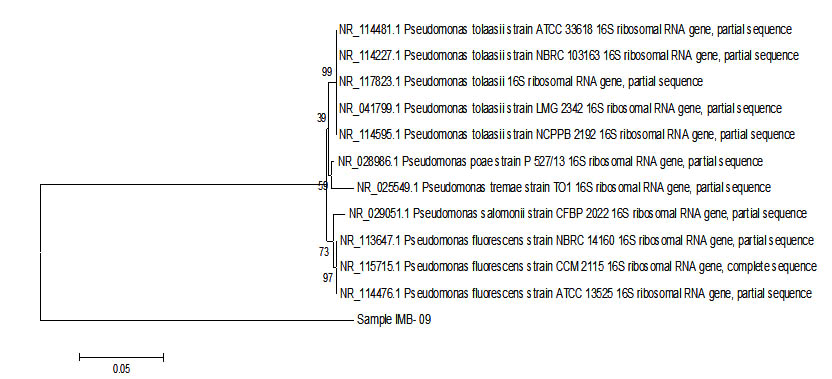
Fig. 3. Phylogenetic tree sample IMB-09 using Mega 6 program.
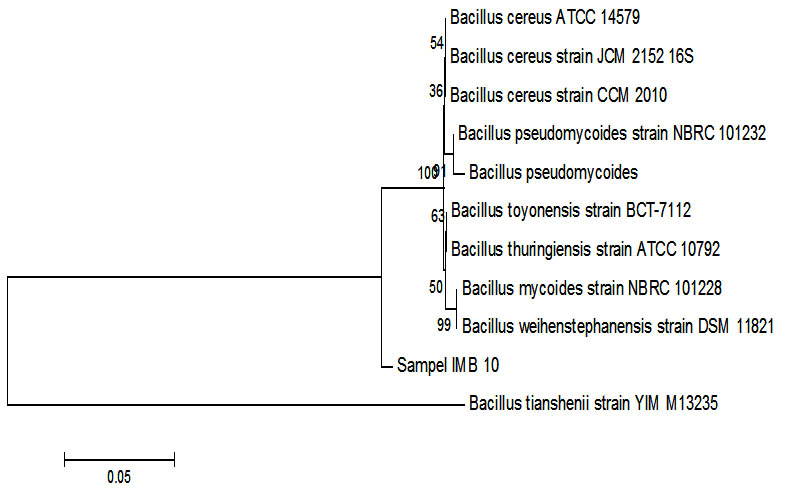
Fig. 4. Phylogenetic tree sample IMB-10 using Mega 6 program.
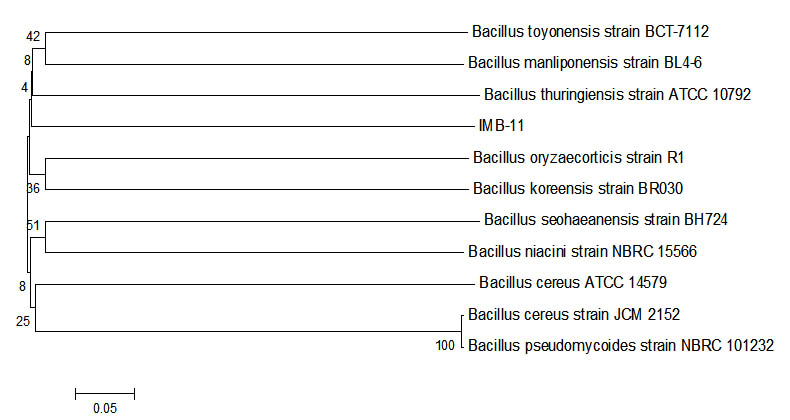
Fig. 5. Phylogenetic tree sample IMB-11 using Mega 6 program.
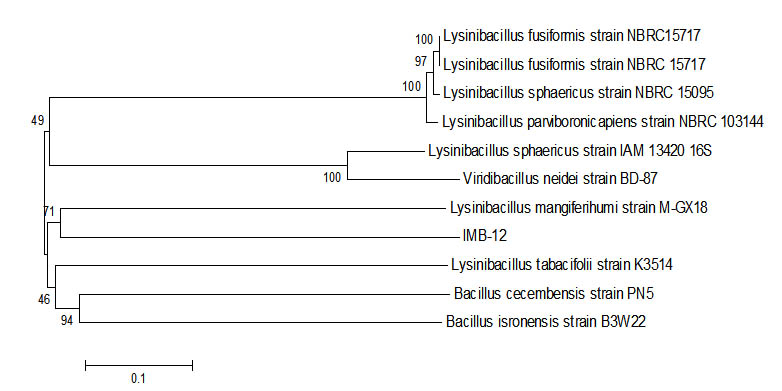
Fig. 6. Phylogenetic tree Sample IMB-12 using Mega 6 program.

Fig. 7. Phylogenetic tree sample IMB-15 using Mega 6 program.
The results of PCR using primer 16s rRNA couples (63F and 1387R) indicates the length of the product PCR obtained i.e. 1,300 BP. Ribbon-Ribbon amplicons result from PCR primer pairs match showed 63F and 1387R against bacterial genome samples are used. The intensity of the PCR product obtained varies in each sample, samples of IMB-9 and IMB-10, this is due to the concentration of the temple used appropriate low, due to the thin insulation results (Figure 10).
Samples IMB-09, IMB-10, IMB-11, IMB-12, IMB and IMB-15 show the results of the nice amplicon and assertive. It also had the same correlations with genomic DNA isolation results obtained as a template on the activities of the PCR.
This bacteria had capability to degrdated coumpound in human body including aromatic hydrocarbon. Inhibition mushroom fungi pathogens7.
Phylogenetic analysis shows that the IMB-10 sample was very similar to the 16s sequencer RNA with Bacillus cereus with value NR 074540.1. Analysis was done using Phylogenetic analysis shows rRNA with Bacillus cereus with value NR 074540.1. Analysis using boosstrap 1,000, this figure states that the sequence stability rate will be estimated use this analysis. Bacillus cereus does have ability to degrade petroleum. The influence of oil contamination the earth in the water will change the response of the membrane bacterial cells thus affecting absorption and the need for bacterial oxygen, so influential to the degradation process8. Bacillus cereus a lot found in wastewater tanks at refinerie soil9.
Baccilus cereus had reported could degradation the phenolic compound in petroleum until 95% and more8. Bacillus cereus strain JMG-01 in enhanced anthracena degradation along the utilization of other hydrocarbons10. Another reported, Bacillus cereus can remove contaminated petroleum in soil or about 30 days11.
IMB 11 had similarities with Bacillus toyonensis strain BCT-7112. Bacillus toyonensis had been reported that it is able to survive in areas that are contaminated by petroleum. Petroleum use as carbon sources12. Analysis of the sequence of the bases of a sample of IMB-11 with all isolates of bacteria from the Blast using a program bio edit and phylogenetic tree analysis using the programe MEGA6. The discovery two isolates that can be degrade petroleum and the two isolates utilized the source petroleum as the only source carbon, as for the two bacterial isolates13. Bacteria degrade petroleum and obtained type of bacteria 94% similarity for Bacillus thuringiensis and Bacillus bombysepticus, and 95% similarity for Bacillus toyonensis BCT-711213,14.
16s rRNA sequence of samples of IMB-12 had similarities with Lysinibacillus fusiform strain NBRC15717obtained from the results of the blast. This bacteria was reported to be used to protect the hulls of the activities of bio fouling17. From the results of research that had been done that the data obtained, a sample of IMB-12 has the ability to degrade petroleum of 62.61%. Data that had been retrieved it states that Lysinibacillus fusiform also had the ability to degrade the oil, which can be applied in the future to handle cases of environmental pollution due to oil spills.
Obtained that samples of IMB-15 had similarities with Pseudomonas stutzeri strain ATCC 17588. Samples of the IMB-15 has the ability to degrade petroleum valued at 76.63%. Pseudomonasstutzeri had been reported to have the ability to degrade petroleum (Celik et al., 2008). These bacteria were also reported to have the same capabilities, it has been reported by (Sazinsky et al., 2004). Researched by Lalucat et al., (2006) this bacteria opportunity pathogen of human and Park et al., (2013) acoording this bacteria have been reported in patients undergoing continuous ambulatory peritoneal dialysis (CAPD).
Over all according to the isolation and identification, bacterias degredation petroleum in waste tank show that bacteria isolate can obtaind nutrien fron hydrocarbon in waste tank via degradation process. This process come from hydrocarbon was degradated by peripheral pathway by oxigenase ensyme to be come a simple compounds like acetile CoA. Five bacterias fond in waste tank can servive because their can degradate hydrocarbon becoming food for life. Microbials comunity had big impect for degrad petroleum.
Fragment of PCR results obtained 16SrRNAs were 1300bp. Blast shown samples of the IMB- 09 was similar to Pseudomonas tollasii, IMB- 10 was similar to Bacillus cereus, IMB-11 was similar to Bacillus toyonensis, IMB-12 was similar to the sample and the fusiform Lysinibacillus fusiformis and IMB-15 Pseudomonas stutzeri. all bacterias have the ability to degrade petroleum.
-
- Chaan, B. Biodegradasi Kimia Organik 2013.http://chaan-biodegradasi-senyawa-organik.html.
- Hajar, D. 2012. Islation, Identification and Isolasi, Identifikasi and Degradation Capability Analysis Hydrocarbon Bacteria Soil Samples B, Cilegon, Banten. [Essay]. Depok. University Indonesia
- Milic, J. S., Beskoski, V. P., Ilic, M. V., Ali, S.A. M., Gojgic-Cvijovic, G. D., and Vrvic, M.M. Bioremediation of soil heavily contaminated with crude oil and its product: composition of the microbial consortium. Journal of the Serbian Chemical Society. 2009; 74(4): 455–460.
- Malatova, K. 2005. Isolation and Characterization of Hydrocarbon Degrading Bacteria from Environmental Habitats In Western New York State Department of Chemistry Rochester Institute of Technology, New York: xi+97hlm.
- Fritsche, W and Hofrichter, M. 2000. Aerobic Degradation by Microorganisms In: H. Rehm and G. Reed (eds). In Biotechnology: Environmental Processes II,Volume11b, Second Edition, Wiley- VCH Verlag GmbH Processes, Germany: 2000; 155pages.
- Hesham Ael L, Mawad AM, Mostafa YM, Shoreit A. 2014. Biodegradation ability and catabolic genes of petroleum degrading Sphingomonas koreensis strain ASU-06 isolated from Egyptian oily soil. Biomed Res Int:127674. doi:10.1155/2014/127674
- Cantore, P. L., Giorgio, A., Lacobellis, N. 2015. Bioactivity of volatile organic compounds produced by Pseudomonas tolaasii. Journal Fronteirs In Microbiology; 6. doi: 10.3389/fmicb. 01082.
- Benerjee, A., Ghosha, A. K . 2016. Biodegradation of Real Petroleum Waste water by Immobilized Hypee Phenol- Tolerant Strain of Bacillus cereus in A Fluidized Bed Bioreactor. Journal Soil Sediment Contamination, 2017; 26(5).
- Agarry S.E., Aremu M.O., Aworanti O.A. 2013. Biodegradation of 2, 6-dichlorophenol wastewater in soil column reactor in the presence of pineapple peels-derived activated carbon, palm kernel oil and inorganic fertilizer.
J. Environt. Protect. 4:537–547. - JDas,M., Bhattacharya, A., Bamu,S., Kotoky, Enhanced Biodegradation of Anthracene by Bacillus cereus Strain JMG-01 Isolated from Hydrocarbon Contaminated Soils. Journal Soil Sediment Contamination, 2017; 26(5).
- Maddela, N.R., Burgos,R., Kadiyala,V., Camon, A. R. Removal of Petroleum Hydrocarbons from Crude Oil In Solid and Slurry Phase By Mixed Soil Microorganisms Isolated From Ecuadorian Oil Fields. Journal International Biodeterioration and Biodegradation. 2016; 108: 85- 90.
- Djamaan, A, Mhd Riza Marjoni, and Friardi Ismed, The Influence of Pretreatment Time, Type and the Concentration of Yeast on Ethanol Production from Rice Straw. Research Journal of Pharmaceutical, Biological and Chemical Sciences, 2015, 6(3): 583-591.
- Gayathri E, Bharathim B, Natarajan S. 2014. Isolation,identification and characterization of diesel degrading bacteria. International Journal of Current Research; 6(6): 7295-300.
- Janssens, T.K.S., Boer, T.E. De., Anamennone, V., Zaagman, N ., Straalen, N.M. V., Roelofs, D. 2016. Draft Genome Sequence Of Bacillus toyonensis VU-DES13, Isolated From Folsomia Candida (Collembola:Entomobrydae). Journal American Society for Microbiology.
- Jiménez N., Morris B. E. L., Cai M., Grnüdger F., Yao J., Richnow H. H. 2012. Evidence for in situ methanogenic oil degradation in the Dagang oil field. Org. Geochem. J. Orggeo Chem : 52 44–54
- Kumar, Arun, and Pradhan Nilotpala. 2013, Inhibition of Pathogenic Bacterial Biofilm by Biosurfactant Produced by Lysinibacillus Fusiformis S9.”doi:10.1007/s00449-013-0976- 5.
- Rivai, H, Asia A, Rina W, Alen Y . Handayani D, Aldi Y, Marlina, and Djamaan A., Isolation of Endophytic Bacteria from Bark, Leaf, and Pericarp of Mangosteen (Garcinia mangostana L.) and Testing of The Antimicrobial Activity, Research Journal of Pharmaceutical, Biological and Chemical Sciences, 2016, 7(1): 1910-1920
- Sazinsky, Matthew H, Joel Bard, Alberto Di Donato ,and Stephen J Lippard, Crystal Structure of the Toluene / O -Xylene Monooxygenase Hydroxylase from Pseudomonas Stutzeri OX1. Journal of Biological Chemistry. 2004; 279(29):
- Lalucat, J., Bennasar, A., Bosch, R.,Valdes, E. G., Palleroni, N. Biology of Pseudomonas stutzeri. Journal Microbiology and Molecular Biology Reviews. 2006; 7(2): 510-547.
- Subhraveti, P., Ong, Q., Keseler, I., Kothari, A., Caspi, R., Karp, P. D. Summary of Bacillus toyonensis, Strain BCT-7112, Version21.5. BioCYC Database Collection 2016.
- Park, S. W., Lee, S. W., Shin, C. H., Kim, G. E., Kim, M, J. Successful Antibiotic Treatment of Pseudomonas stutzeri Induced Peritoritis without Peritoneal Dialysis Catheter Removal In Countinous Ambulatory Peritoneal Dialysis. Journal Kidney Research and Clinical Practice. 2013; 32(2): 81- 83.
- Zam, S .I. Bioremediation of Petroleum Refining Waste Pertamina UP II Pakning River by Using Indigenous Bacteria. Thesis. Bandung Institute of Technology. Bandung, 2006
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


