ISSN: 0973-7510
E-ISSN: 2581-690X
In this study, the isolation of lactic acid bacteria was carried out from one hundred white cheese samples collected from different regions of Turkey. Subsequently, phenotypic and genotypic characterization of the isolates was performed. Biochemical characteristics of the isolates were determined by API 50CHL. Furthermore, the biotechnological enzyme production potential of the isolates was screened. Genomic fingerprint profiles of the test isolates were detected by using rep-PCR (BOX-PCR), which has been used successfully in the differentiation of microorganisms at the species, subspecies, and even strain levels. The results showed that a total of forty-one bacteria were isolated and seventeen of which are found to be different species. The isolates generally grew at 4-6 pH values, 0-8% NaCl and 30-40°C. Later, isolates thought to be different species were identified by 16S rRNA gene sequence analyses. According to 16S rRNA sequence results, MA56 showed a 96.41% similarity match to Lentilactobacillus buchneri, it is thought to be a new species. In addition, MA19, MA25, MA43, and MA47 were determined to have multi-enzyme production potential. MA43 has a plantaricin gene and it showed a high antagonistic effect on Escherichia coli O157:H7 ATCC 43888 and Pseudomonas aeruginosa ATCC 9027. Inhibition zones were measured at 19 mm and 16 mm respectively.
Lactic Acid Bacteria, Bacteriocin, Molecular Characterization, Biotechnological Enzymes
Cheese is produced by coagulating the milk with the effect of a suitable proteolytic enzyme or organic acids.1,2 The unique taste and aroma of cheeses are obtained by the addition of lactic acid bacteria to the culture. Lactic acid bacteria (LAB) are gram-positive bacteria that produce lactic acid as the main product during the fermentation of carbohydrates.3-5 They are non-spore-forming, anaerobic or microaerophilic, and acid-tolerant organisms with a rod or coccal cellular shapes.6,7 LAB is a group of bacteria that consists of the genus including Streptococcus, Lactococcus, Pediococcus, Enterococcus, and Lactobacillus which is commonly found in dairy and fermented foods.8-11 Many studies are reporting the health benefits of fermented dairy products. Fermented foods typically contain microorganisms considered to be Generally Regarded As Safe (GRAS), which can produce a range of beneficial by-products/metabolites such as antimicrobial peptides (e.g. bacteriocins), ethanol, organic acids, fatty acids, and carbon dioxide.12-14 It is known that products resulting from LAB-induced fermentations have anti-cancer, immunomodulatory,15 anti-gastritis,16 antihypertensive,17 and anti-allergenic effects.18 In addition, Mozaffarian et al reported that consuming LAB fermented foods had positive effects on body fitness.19,20 Other studies have shown that the consumption of fermented yogurt and dairy products might reduce the risk of developing cardiovascular disease (CVD) and type 2 diabetes mellitus (T2DM).21,22 In addition, researchers reported that fermented milk and dairy products associated with LAB have hypocholesteremic23 and anti-cancer properties.24
Besides the health aspect, LABs can also be the source of new species with enzymatic activities for biotechnological properties.25 Microbial enzymes are more preferred than other enzymes because they have high catalytic activity and efficiency. Amylases, proteases, and lipases are commonly used in biotechnological processes. LAB with amylase, lipase, xylanases, and protease activities have been reported in previous studies.26-29 Considering these properties, interest in lactic acid bacteria is increasing day by day and there are many LAB species with biotechnological potential yet to be discovered. In this study, isolation, identification, and molecular characterization of lactic acid bacteria from cheese samples collected from different regions (Erzurum, Van, Konya, Karaman, and Kars) of Turkey. Later, biotechnologically important enzyme and bacteriocin production potentials of the isolates were determined.
Sampling and lactic acid bacteria isolation
A total of one hundred cow cheese samples taken from markets in different regions of Turkey (Erzurum, Van, Konya, Karaman, and Kars) were brought to the laboratory under aseptic conditions and kept at + 4°C until use. 225 ml of sterile physiological water (0.9% NaCl) was added to a 25g cheese sample and homogenization process was carried out and dilution series (100-10-7) were prepared.30 The dilution aliquots were spread on MRS and M17 Agar media and incubated at 35°C for 48 hours. Pure cultures of isolates were obtained and stored at -86°C in a stock medium containing 15% glycerol.31
Phenotypic Characterization
To determine phenotypic characterization, test strains were grown in MRS and M17 medium at different temperatures 15°C to 50°C (with 5°C intervals) for up to 72 h. The pH ranges were analyzed in MRS and M17 at pH 3.0–11.0 (1 pH unit intervals), and tolerance of NaCl was determined using MRS and M17 supplemented with 0–12 % NaCl (at intervals of 1.0 %) for 72 h. All experiments were tested in triplicate and growth was measured at OD600 nm.32 Gram-staining of test strains was performed according to the method by Gerhardt et al.33 Catalase activity was performed by the production of bubbles of a drop of 3 % H2O2 (v/v). Oxidase reagent (Sigma) was used for testing oxidase activity.34 Furthermore, biochemical characterization of isolates was conducted using API 50CHL test.35
Genotypic Characterization
Genomic DNA isolation was performed according to the WizardR Genomic DNA Purification Kit (Promega, Southampton, UK, A2360) protocol. The rep-PCR reactions were carried out in a Sensequest Thermal Cycler (Göttingen, Germany) using BOXAIR primer (5′-CTACGGCAAGGCGACGCTGACG-3′), PCR mixtures contained: 5 μl Gitschier Buffer, 12.7 μl ddH2O, 2.5 μl dimethyl sulfoxide, 1.25 μl bovine serum albumin, 1.25 μl dNTP, 4 μl primer, 0.3 μl Taq DNA polymerase and 3 μl template DNA. PCR Cycles were, initial denaturation at 94°C for 7 min., 36 cycles of 1 min. at 94°C, 1 min. at 45°C, 8 min. at 65°C. Final extension at 65°C for 16 min. At the end of PCR, samples were run in 1% agarose gel for 90 minutes.36
16S rRNA region was amplified using 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) primers37. 30 μL volume of PCR mixture containing, 13.1 µl ddH2O, 3 µl 10X PCR buffer, 1.8 µl MgCl2, 1.2 µl DMSO, 0.6 µl dNTP, 3 µl (5 µM) forward primer (27F), 3 µl (5µM) reverse primer (1492R), 0.3 µl Taq DNA polymerase and 4 µl template DNA (70 ng). The amplified fragments were cloned into Escherichia coli JM101 strain with the pGEM-T Easy Cloning Vector (Promega, Southampton, UK) according to the instructions of the manufacturer. After the cloning, plasmid isolation was performed by selecting colonies that gave the positive result, and the sequence analysis was made by the Macrogen Company (Netherlands). The 16S rRNA obtained was compared with other bacterial series in GenBank and EzTaxon (http://blast.ncbi.nlm.nih and http://www.eztaxon.org), the similarity rate between them was determined and GenBank accession numbers were received.38,39
Preliminary enzyme assays
Lipase
To test lipase enzyme production, isolates were inoculated into a tributyrin agar medium containing 1 % tributyrin (glycerol tributyrate) and incubated at 35°C for 48 h. Isolates with a clear zone were considered positive for lipase.40,41
Amylase
Test strains were streaked on plates containing MRS (de Man, Rogosa and Sharpe) agar with 1% starch instead of glucose that were incubated at 35°C for 48 h. Then, Petri dishes were treated with Lugol’s solution. Isolates showing clear zones were evaluated as amylase positive.42
Protease
To test whether isolates can produce protease enzymes, the strain was inoculated into MRS agar containing 1% Skimmed Milk Powder and incubated at 35°C for 48 h. Isolates with a clear zone were considered positive for protease.40
Xylanase
Isolates were inoculated into medium containing xylan (10 g/L), NaNO3 (1.2 g/L), KH2PO4 (13 g/L), K2HPO4 (6 g/L), CaCl2 (0.05 g/L), MgSO4 (0.01 g/L ), ZnSO4 (0.001 g/L) and agar (15 g/L) incubated at 35°C for 48 hours. After incubation, the plates were stained with 0.1% congo red for 20 min and then washed with 1M NaCl. Isolates with orange-colored zones were evaluated for xylanase positive.29
Bacteriocin gene detection
After the characterization of test strains, bacteriocin production characteristics of isolates were investigated. For this purpose, PCR analysis was performed using primers specific to each bacteriocin gene.43,44 The 16S rRNA PCR program given above was performed except for changing the annealing temperatures of bacteriocin primers.
Detection of Antibacterial Activity
For the detection of antibacterial activity, a disc diffusion assay was used. Pathogenic bacteria were spread on the surfaces of Mueller Hinton agar media. Overnight cultures, on MRS medium, of the strains to be tested were centrifuged and cell-free supernatant was loaded on discs placed in the middle of the petri dish. The plates were incubated at 37°C for 24h. The antagonistic effects of the test strains were determined by measuring the zone of inhibition diameters.45 The target test strains used in this study were Escherichia coli O157:H7 (ATCC 43888), Salmonella typhimurium (ATCC 14028), Serratia marcescens (ATCC 810), Pseudomonas aeruginosa (ATCC 9027), Streptococcus pyogenes (ATCC 12344), Klebsiella pneumoniae (ATCC 13883), Listeria monocytogenes (ATCC 7644), Staphylococcus epidermidis (ATCC 12228), Shigella dysenteriae (ATCC 13313) and Staphylococcus aureus (ATCC 6538).
Isolation of LAB
In this study, a total of 41 bacterial isolates were isolated from cheese samples taken from provinces (Erzurum, Kars, Karaman, Konya, and Van). Since seventeen of the isolated bacteria belonged to different species (according to rep-PCR results), stock cultures of these strains were prepared and the study was continued with these isolates.
Phenotypic Characterization
According to conventional analysis, all isolates were gram-positive and oxidase negative and showed cocci or bacilli cell morphology. In general isolates grow at 4-6 pH values, 0-8% NaCl and 30-40°C. Similarly, Ni et al determined that isolated lactic acid bacteria can usually grow at 35-45°C at pH 3 and 6.5% NaCl.46 Interestingly, it has been found that the MA7 strain can thrive in a wide range of pH and salt concentrations such as pH:2-11 and 0-10% NaCl. So, MA7 can be suitable for many biotechnological processes. Detailed phenotypic characteristics and API test results are given in Table 1.
Table (1):
Phenotypic characterization of isolates 1: MA4, 2: MA7, 3: MA10, 4: MA12, 5: MA19, 6: MA25, 7: MA 27, 8: MA28, 9: MA31, 10: MA33, 11: MA34, 12: MA35, 13: MA39, 14: MA43, 15: MA47, 16: MA55, 17: MA56
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
pH |
4-6 |
2-11 |
4-6 |
2-8 |
3-6 |
4-6 |
4-7 |
4-5 |
4-6 |
4-6 |
4-6 |
4-6 |
5-7 |
4-6 |
3-6 |
4-6 |
2-8 |
NaCl |
0-9 |
0-10 |
0-9 |
0-8 |
4-6 |
0-6 |
0-8 |
0-11 |
0-8 |
0-6 |
0-8 |
0-8 |
0-8 |
0-5 |
0-8 |
0-10 |
0-8 |
Temperature |
30-40 |
30-40 |
15-45 |
30-40 |
30-40 |
30-40 |
15-45 |
15-45 |
15-45 |
30-40 |
30-40 |
15-45 |
30-40 |
30-40 |
30-40 |
30-40 |
30-40 |
Morphology |
Bacil |
Bacil |
Bacil |
Bacil |
Coc |
Coc |
Coc |
Bacil |
Coc |
Coc |
Bacil |
Coc |
Coc |
Bacil |
Coc |
Coc |
Bacil |
Oxidase |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
Catalase |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
L-Arabinose |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
– |
+ |
Ribose |
– |
+ |
+ |
– |
– |
– |
+ |
+ |
d |
– |
+ |
+ |
– |
+ |
– |
– |
+ |
Xylose |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
– |
– |
– |
– |
+ |
Adonitol |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
Galactose |
– |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
– |
+ |
+ |
D-Glucose |
– |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
– |
+ |
+ |
D-Fructose |
– |
+ |
+ |
– |
– |
– |
+ |
+ |
– |
+ |
+ |
+ |
+ |
+ |
– |
+ |
+ |
D-Mannose |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
d |
+ |
– |
+ |
– |
Rhamnose |
– |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
d |
– |
– |
– |
– |
– |
Dulcitol |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
– |
– |
– |
Mannitol |
– |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
+ |
– |
– |
– |
Sorbitol |
– |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
– |
– |
– |
Met-D-Gluc. |
– |
– |
+ |
– |
– |
– |
– |
d |
– |
– |
– |
– |
– |
– |
– |
– |
+ |
NAG |
– |
+ |
+ |
– |
– |
– |
+ |
d |
d |
+ |
+ |
+ |
– |
+ |
– |
+ |
+ |
Amygdaline |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
Arbutine |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
+ |
– |
– |
+ |
– |
– |
– |
Esculine |
d |
+ |
– |
+ |
– |
– |
+ |
– |
– |
– |
– |
+ |
– |
+ |
– |
+ |
– |
Salicine |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
+ |
d |
d |
+ |
– |
d |
– |
Cellobiose |
– |
+ |
+ |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
– |
+ |
– |
+ |
– |
Maltose |
– |
d |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
+ |
– |
+ |
+ |
– |
– |
+ |
Lactose |
– |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
– |
+ |
+ |
+ |
– |
+ |
+ |
Melibiose |
– |
– |
– |
– |
+ |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
Saccharose |
– |
d |
+ |
– |
– |
– |
– |
– |
+ |
d |
– |
– |
+ |
+ |
– |
– |
– |
Trehalose |
– |
+ |
+ |
– |
– |
– |
+ |
– |
+ |
+ |
+ |
+ |
+ |
+ |
– |
d |
+ |
Melezitose |
– |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
d |
+ |
– |
– |
– |
D-Raffinose |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
+ |
Gentiobiose |
– |
– |
+ |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
– |
+ |
– |
+ |
– |
D-Turanose |
– |
+ |
– |
– |
– |
– |
– |
– |
d |
d |
+ |
– |
d |
– |
– |
– |
– |
D-Tagatose |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
+ |
d |
– |
– |
– |
+ |
– |
D-Arabitol |
– |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
d |
– |
– |
– |
L-Arabitol |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
+:Positive -: Negative d: Delayed
Genotypic Characterization
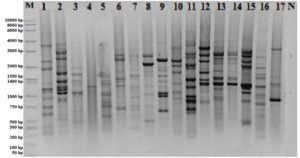
Previous studies have reported that rep-PCR is an easy method that can be used to classify bacteria. Mohammed et al used BOX-PCR analysis to characterize lactic acid bacteria isolated from traditional milk samples.47 We also performed genomic fingerprint analysis of isolates using BOX-PCR in this study. While 12 polymorphic bands were observed in some of the test strains, it was observed that there was 1 band in some isolates (Figure 1). It was observed that the BOX-PCR was not sufficient to classify all LAB.
Figure 1. BOX-PCR profiles of isolates ( M:Marker, 1: MA4, 2: MA7, 3: MA10, 4: MA12, 5: MA19, 6: MA25, 7: MA27, 8: MA28, 9: MA31, 10: MA33, 11: MA34, 12: MA35, 13: MA39, 14: MA43, 15: MA47, 16: MA55, 17:MA56, N: Negative Control)
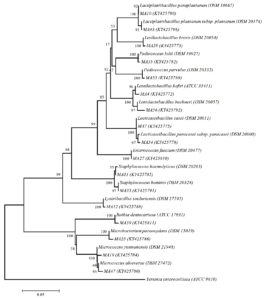
16S rRNA sequence analysis is used as a powerful tool in determining the prokaryotic diversity in almost every environment.48,49 So, we performed molecular identification of lactic acid bacteria isolated from cheese samples based on 16S rRNA sequencing. Except for MA56, all isolates were found to be 99% similar to related standard type strains. MA56 has a 96.41% similarity match with Lentilactobacillus buchneri DSM 20057. According to general acceptance, the 16S rRNA gene sequence similarity ratio below 97% is a new species indicator of the isolate.49 In recent years, it has been reported that this rate is 98.2% – 99% and that the whole genome sequence and DNA: DNA hybridization is required in addition to the 16S rRNA gene sequence.50 According to this information, MA56 might be a novel species belonging to the genus Lentilactobacillus. It is thought that MA56 will be added to the literature as a novel species as a result of the whole genome sequence analysis in future studies. Detailed sequence results of the isolates and related species are given in Table 2. Also, the 16S rRNA gene-based phylogenetic tree is shown in Figure 2.
Table (2):
Related species and similarity rates of isolates according to 16S rRNA sequence analysis.
Isolate Code |
Product size (bp) |
Related species |
Similarity rate (%) |
Genbank No. |
|---|---|---|---|---|
MA 4 |
1535 |
Lentilactobacillus kefiri |
99 |
KY425772 |
MA 7 |
813 |
Lacticaseibacillus casei |
99 |
KY425775 |
MA 10 |
1528 |
Lactiplantibacillus paraplantarum |
99 |
KY425790 |
MA 12 |
1515 |
Lysinibacillus sindurienssis |
99 |
KY425788 |
MA 19 |
1484 |
Micrococcus yunnanensis |
99 |
KY425784 |
MA 25 |
1486 |
Microbacterium paraoxydans |
99 |
KY425786 |
MA 27 |
1387 |
Enterococcus faecium |
99 |
KY425810 |
MA 28 |
1527 |
Levilactobacillus brevis |
99 |
KY425773 |
MA 31 |
1512 |
Staphylococcus haemolyticus |
99 |
KY425785 |
MA 33 |
885 |
Staphylococcus hominis |
99 |
KY425791 |
MA 34 |
1531 |
Lacticaseibacillus paracasei subsp. paracesi |
99 |
KY425778 |
MA 35 |
1538 |
Pediococcus lolii |
99 |
KY425782 |
MA 39 |
1488 |
Rothia dentocariosa |
99 |
KY425811 |
MA 43 |
1528 |
Lactiplantibacillus plantarum subsp. plantarum |
99 |
KY425796 |
MA 47 |
1462 |
Micrococcus aloeverae |
99 |
KY425780 |
MA 55 |
1539 |
Pediococcus parvulus |
99 |
KY425789 |
MA 56 |
1491 |
Lentilactobacillus buchneri |
96 |
KY425792 |
Figure 2. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences of test strains and related type species. Yersinia enterocolitica ATCC 9610 was used as an out-group. Bootstrap values based on 1000 replications are listed as percentages at branching points. The accession numbers are given in parentheses. The scale bar represented 0.5% divergence
Determination of Biotechnological Enzyme Production Characteristics of Isolates
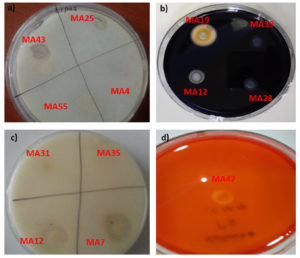
The ability of isolates to produce amylase, lipase, protease and xylanase enzymes, which are biotechnologically important, was determined. As a result of the analysis, one strain has xylanase and lipase activity, five strains with amylase activity, and ten strains with protease activity were observed (Table 3). Also, Petri images of some isolates are given in Figure 3. Matthews et al investigated the enzyme production potential of lactic acid bacteria and determined that especially Lactobacillus and Pediococcus species are important producers of lipase, cellulase, and xylanase enzymes. In another study, Konkit and Kim examined that Lactoccocus chungangensis produces amylase, proteinase, and lipase enzymes.26 These enzymes are very important for industrial processes. For example, lipases are used for transesterification acidolysis, xylanase is used for the enzymatic breakdown of agricultural wastes for the production of alcohol fuels, enzymatic treatment of animal feed to release free pentose sugars, manufacturing of dissolving pulps yielding cellulose for rayon production, and bio-bleaching of wood pulps, proteases used for the detergent industry, and amylases used for food, fermentation and pharmaceutical industries.
Table (3):
Screening of Industrial Enzyme Profiles of Isolates.
Isolate Code |
Related species |
Amylase |
Lipase |
Protease |
Xylanase |
|---|---|---|---|---|---|
MA4 |
Lentilactobacillus kefiri |
– |
– |
++ |
– |
MA7 |
Lacticaseibacillus casei |
– |
– |
++ |
– |
MA10 |
Lactiplantibacillus paraplantarum |
– |
– |
– |
– |
MA12 |
Lysinibacillus sinduriensis |
– |
– |
++ |
– |
MA19 |
Micrococcus yunnanensis |
++ |
– |
++ |
– |
MA25 |
Microbacterium paraoxydans |
+ |
+ |
+ |
– |
MA28 |
Levilactobacillus brevis |
– |
– |
++ |
– |
MA31 |
Staphylococcus haemolyticus |
– |
– |
– |
– |
MA33 |
Staphylococcus hominis |
– |
– |
++ |
– |
MA34 |
Lactiplantibacillus paracasei subsp. paracesi |
– |
– |
– |
– |
MA35 |
Pediococcus lolii |
+ |
– |
– |
– |
MA39 |
Rothia dentocariosa |
– |
– |
– |
– |
MA43 |
Lactiplantibacillus plantarum subsp. plantarum |
+ |
+ |
++ |
– |
MA47 |
Micrococcus aloeverae |
– |
– |
++ |
+ |
MA55 |
Pediococcus parvulus |
+ |
– |
– |
– |
MA56 |
Lentilactobacillus buchneri |
– |
– |
++ |
– |
-: Negative, +: Positive, ++: Strong positive
Figure 3. Screening biotechnological enzymes profiles of some isolates a) Petri image of lipase b) Petri image of amylase c) Petri image of protease d) Petri image of xylanase
There are many studies in the literature in which enzymes obtained from lactic acid bacteria are used in biotechnological processes. For example, The xylanase enzyme purified from Pedioccus acidilactici was applied in clarification of fruit juices.29 In another study, it was reported that the protease enzyme obtained from Lactobacillus plantarum had an antimicrobial effect on pathogenic microorganisms.51 Therefore, in this study, isolates coded MA19, MA25, MA43, and MA47 were determined to have multi-enzyme production potential (Table 3). These isolates are attractive for biotechnological processes because they have more than one enzyme activity.
Determination of Bacteriocin Production Potential
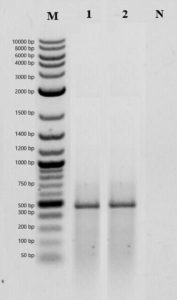
The presence of bacteriocin genes in the strains was determined using bacteriocin-specific primers with PCR. As a result of the PCR analysis, it was determined that only the MA43 strain has a plantaricin gene region. The gel image of plantaricin belonging to MA43 and Lactobacillus plantarum ATCC 8014 are shown in Figure 4.
Figure 4. PCR analysis result of plantaricin genes. M: Marker, 1: Lactobacillus plantarum ATCC 8014, 2: MA43, 3: Negative control
After it was determined that MA43 has a bacteriocin gene, its effect on pathogenic bacteria was investigated. It has been determined that MA43 has the highest antibacterial effect against Escherichia coli O157:H7 ATCC 43888 and Pseudomonas aeruginosa ATCC 9027 (Table 4). Previous studies have also reported that Lactobacillus plantarum has an antimicrobial effect against various pathogens.52,53 In this respect, MA43 has an antagonistic effect against foodborne pathogens showing that it can be used as a food preservative.
Table (4):
The result of the disc diffusion test. The diameter of the inhibition zone of MA43.
Pathogens |
MA43 |
|---|---|
Serratia marcescens ATCC 810 |
5 ± 0.4 |
Shigella dysenteriae ATCC 13313 |
12 ± 1.3 |
Klebsiella pneumoniae ATCC 13883 |
12 ± 0.8 |
Streptococcus pyogenes ATCC 12344 |
10 ± 0.4 |
Staphylococcus epidermidis ATCC 12228 |
9 ± 0.3 |
Staphylococcus aureus ATCC 6538 |
12 ± 0.8 |
Pseudomonas aeruginosa ATCC 9027 |
16 ± 1.1 |
Salmonella typhimurium ATCC 14028 |
7 ± 0.6 |
Listeria monocytogenes ATCC 7644 |
9 ± 0.7 |
Escherichia coli O157:H7 ATCC 43888 |
19 ± 0.5 |
The measures of the inhibition zone are expressed in mm.
In this study, it was determined that the white cheese samples have a very wide range of microflora. 16S rRNA sequence similarity to the closest species of the MA56 was determined as 96.41%. This isolate is more likely to be a novel species of lactic acid bacteria. MA43 not only has amylase, lipase, and protease activity but also produces bacteriocin making it unique for biotechnological processes. In addition, this study will be a pioneer for future studies to be conducted on MA56 and MA43.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This research was supported by Ataturk University, project no. FBG-2020-8779.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Domingos-Lopes MFP, Stanton C, Ross PR, Dapkevicius MLE, Silva CCG. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017;63:178-190.
Crossref - Carafa I, Nardin T, Larcher R, Viola R, Tuohy K, Franciosi E. Identification and characterization of wild lactobacilli and pediococci from spontaneously fermented Mountain cheese. Food Microbiol. 2015;48:123-132.
Crossref - Alvarez-Sieiro P, Montalban-Lopez M, Mu DD, Kuipers OP. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100(7):2939-2951.
Crossref - Varoquaux P, Wiley RC. Biological and Biochemical Changes in Minimally Processed Refrigerated Fruits and Vegetables. Minimally Processed Refrigerated Fruits and Vegetables, 2nd Edition 2017:153-186.
Crossref - Leong KH, Chen YS, Pan SF, et al. Diversity of Lactic Acid Bacteria Associated with Fresh Coffee Cherries in Taiwan. Curr Microbiol. 2014;68(4):440-447.
Crossref - Agriopoulou S, Stamatelopoulou E, Sachadyn-Krol M, Varzakas T. Lactic Acid Bacteria as Antibacterial Agents to Extend the Shelf Life of Fresh and Minimally Processed Fruits and Vegetables: Quality and Safety Aspects. Microorganisms. 2020;8(6):952.
Crossref - Shoukat S. Potential anti-carcinogenic effect of probiotic and lactic acid bacteria in detoxification of benzo[a]pyrene: A review. Trends Food Sci Technol. 2020;99:450-459.
Crossref - Barbieri F, Montanari C, Gardini F, Tabanelli G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods. 2019;8(1):17.
Crossref - Azam M, Mohsin M, Ijaz H, et al. Lactic acid bacteria in traditional fermented Asian foods. Pak J Pharm Sci. 2017;30:1803-1814.
- von Mollendorff JW, Todorov SD, Dicks LMT. Comparison of bacteriocins produced by lactic-acid bacteria isolated from boza, a cereal-based fermented beverage from the Balkan Peninsula. Curr Microbiol. 2006;53(3):209-216.
Crossref - Szutowska J, Gwiazdowska D. Probiotic potential of lactic acid bacteria obtained from fermented curly kale juice. Arch Microbiol. 2021;203(3):975-988.
Crossref - Marco ML, Heeney D, Binda S, et al. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol. 2017;44:94-102.
Crossref - Macori G, Cotter PD. Novel insights into the microbiology of fermented dairy foods. Curr Opin Biotechnol. 2018;49:172-178.
Crossref - Bindu A, Lakshmidevi N. Identification and in vitro evaluation of probiotic attributes of lactic acid bacteria isolated from fermented food sources. Arch Microbiol. 2021;203(2):579-595.
Crossref - Qian BJ, Xing MZ, Cui L, et al. Antioxidant, antihypertensive, and immunomodulatory activities of peptide fractions from fermented skim milk with Lactobacillus delbrueckii ssp bulgaricus LB340. J Dairy Res. 2011;78(1):72-79.
Crossref - Rodriguez C, Medici M, Rodriguez AV, Mozzi F, Valdez GF. Prevention of chronic gastritis by fermented milks made with exopolysaccharide-producing Streptococcus thermophilus strains. J Dairy Sci. 2009;92(6):2423-2434.
Crossref - Mallappa RH, Balasubramaniam C, Nataraj BH, et al. Microbial diversity and functionality of traditional fermented milk products of India: Current scenario and future perspectives. Int Dairy J. 2021;114:104941.
Crossref - Drywien M, Frackiewicz J, Gornicka M, Gadek J, Jalosinska M. Effect of probiotic and storage time of thiamine and riboflavin content in the milk drinks fermented by Lactobacillus casei KNE-1. Rocz Panstw Zakl Hig. 2015;66:373-377.
- Mozaffarian D, Hao T, Rimm EB, Willett WC, Hu FB. Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med. 2011;364(25):2392-2404.
Crossref - Chen M, Sun Q, Giovannucci E, et al. Dairy consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. BMC Med. 2014;12:215.
Crossref - Eussen SJ, van Dongen MC, Wijckmans N, et al. Consumption of dairy foods in relation to impaired glucose metabolism and type 2 diabetes mellitus: the Maastricht Study. Br J Nutr. 2016;115(8):1453-1461.
Crossref - Soedamah-Muthu SS, Masset G, Verberne L, Geleijnse JM, Brunner EJ. Consumption of dairy products and associations with incident diabetes, CHD and mortality in the Whitehall II study. Br J Nutr. 2013;109(4):718-726.
Crossref - Lye HS, Rahmat-Ali GR, Liong MT. Mechanisms of cholesterol removal by lactobacilli under conditions that mimic the human gastrointestinal tract. Int Dairy J. 2010;20(3):169-175.
Crossref - Kapila S, Sinha PR, Singh S. Influence of feeding fermented milk and non-fermented milk containing Lactobacillus casei on immune response in mice. Food Agric Immunol. 2007;18(1):75-82.
Crossref - Mathur H, Beresford TP, Cotter PD. Health Benefits of Lactic Acid Bacteria (LAB) Fermentates. Nutrients. 2020;12(6):1679.
Crossref - Konkit M, Kim W. Activities of amylase, proteinase, and lipase enzymes from Lactococcus chungangensis and its application in dairy products. J Dairy Sci. 2016;99(7):4999-5007.
Crossref - Unban K, Kanpiengjai A, Takata G, Uechi K, Lee W-C, Khanongnuch C. Amylolytic Enzymes Acquired from L-Lactic Acid Producing Enterococcus faecium K-1 and Improvement of Direct Lactic Acid Production from Cassava Starch. Appl Biochem Biotechnol. 2017;183(1):155-170.
Crossref - Saez GD, Saavedra L, Hebert EM, Zarate G. Identification and biotechnological characterization of lactic acid bacteria isolated from chickpea sourdough in northwestern Argentina. Lwt-Food Science and Technology. 2018;93:249-256.
Crossref - Adiguzel G, Faiz O, Sisecioglu M, et al. A novel endo-beta-1,4-xylanase from Pediococcus acidilactici GC25; purification, characterization and application in clarification of fruit juices. Int J Biol Macromol. 2019;129:571-578.
Crossref - Luiz LMP, Castro RD, Sandes SHC, et al. Isolation and identification of lactic acid bacteria from Brazilian Minas artisanal cheese. CyTA-Journal of Food. 2017;15(1):125-128.
Crossref - Kirmaci HA, Ozer BH, Akcelik M, Akcelik N. Identification and characterisation of lactic acid bacteria isolated from traditional Urfa cheese. Int J Dairy Technol. 2016;69(2):301-307.
Crossref - Baltaci MO, Ay H, Akbulut S, et al. Bacillus pasinlerensis sp. nov., a thermophilic bacterium isolated from a hot spring in Turkey. Int J Syst Evol Microbiol. 2020;70(6):3865-3871.
Crossref - Gerhardt P, Murray R, Costilow RN, et al. Manual of Methods for General Bacteriology. 1981.
- Adiguzel A, Ay H, Baltaci MO, Akbulut S, Albayrak S, Omeroglu MA. Genome-based classification of Calidifontibacillus erzurumensis gen. nov., sp. nov., isolated from a hot spring in Turkey, with reclassification of Bacillus azotoformans as Calidifontibacillus azotoformans comb. nov. and Bacillus oryziterrae as Calidifontibacillus oryziterrae comb. nov. Int J Syst Evol Microbiol. 2020;70(12):6418-6427.
Crossref - Anekella K, Perez-Diaz IM. Characterization of robust Lactobacillus plantarum and Lactobacillus pentosus starter cultures for environmentally friendly low-salt cucumber fermentations. J Food Sci. 2020;85(10):3487-3497.
Crossref - Saez GD, Hebert EM, Saavedra L,Zarate G. Molecular identification and technological characterization of lactic acid bacteria isolated from fermented kidney beans flours (Phaseolus vulgaris L. and P. coccineus) in northwestern Argentina. Food Research International. 2017;102:605-615.
Crossref - Baltaci MO, Adiguzel A. Isolation, Identification and Molecular Characterization of Cellulolytic Bacteria from Rumen Samples collected from Erzurum Slaughter House, Turkey. Res J Biotechnol. 2016;11:32-38.
- Baltaci MO, Genc B, Arslan S, Adiguzel G, Adiguzel A. Isolation and Characterization of Thermophilic Bacteria from Geothermal Areas in Turkey and Preliminary Research on Biotechnologically Important Enzyme Production. Geomicrobiol J. 2017;34(1):53-62.
Crossref - Muslu S, Genc B, Adiguzel M, Seyda A, Ahmet A. Proteolytic, Lipolytic and Amylolytic Bacteria Reservoir of Turkey; Cold-Adaptive Bacteria in Detergent Industry. J Pure Appl Microbiol. 2020;14(1):63-72.
Crossref - Linares-Morales JR, Cuellar-Nevarez GE, Rivera-Chavira BE, Gutierrez-Mendez N,Perez-Vega SB, Nevarez-Moorillon GV. Selection of Lactic Acid Bacteria Isolated from Fresh Fruits and Vegetables Based on Their Antimicrobial and Enzymatic Activities. Foods. 2020;9(10):1399.
Crossref - Albayrak S, Genc B, Ozkan H, Mesut T, Ahmet A. Presence of Different Bacterial Species in Thermal Sources and Novelty in Their Industrial Enzyme Productions. J Pure Appl Microbiol. 2019;13(3):1375-1387.
Crossref - Padmavathi T, Bhargavi R, Priyanka PR, Niranjan NR, Pavitra PV. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J Genet Eng Biotechnol. 2018;16(2):357-362.
Crossref - Chiorean S, Vederas JC, van Belkum MJ. Identification and Heterologous Expression of the sec-Dependent Bacteriocin Faerocin MK from Enterococcus faecium M3K31. Probiotics and Antimicrobial Proteins. 2018;10(2):142-147.
Crossref - Erkaya E, Genc B, Akbulut S, et al. Bacteriocin Producing Bacteria Isolated from Turkish Traditional Sausage Samples. J Pure Appl Microbiol. 2020;14(2):1567-1576.
Crossref - Mezaini A, Chihib NE, Bouras AD, Nedjar-Arroume N, Hornez JP. Antibacterial activity of some lactic acid bacteria isolated from an Algerian dairy product. J Environ Public Health. 2009;2009:678495.
Crossref - Ni K, Wang Y, Li D, Cai Y, Pang H. Characterization, Identification and Application of Lactic Acid Bacteria Isolated from Forage Paddy Rice Silage. Plos One. 2015;10(3):e0121967.
Crossref - Mohammed M, Abd El-Aziz H, Omran N, El-Soda SAM. Rep-PCR characterization and biochemical selection of lactic acid bacteria isolated from the Delta area of Egypt. Int J Food Microbiol. 2009;128(3):417-423.
Crossref - Martinez-Porchas M, Villalpando-Canchola E, Ortiz Suarez LE, Vargas-Albores F. How conserved are the conserved 16S-rRNA regions? Peer J .2017;5:e3036.
Crossref - Edgar RC. Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. Peer J. 2018;6:e4652.
Crossref - Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018;34:2371-2375.
Crossref - Lin TH, Pan TM. Characterization of an antimicrobial substance produced by Lactobacillus plantarum NTU 102. J Microbiol Immunol Infect. 2019;52(3):409-417.
Crossref - Wang S, Peng Q, Jia HM, et al. Prevention of Escherichia coli infection in broiler chickens with Lactobacillus plantarum B1. Poultry Science. 2017;96(8):2576-2586.
Crossref - Layus BI, Gerez CL, Rodriguez AV. Antibacterial Activity of Lactobacillus plantarum CRL 759 Against Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa. Arab J Sci Eng. 2020;45:4503-4510.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.