ISSN: 0973-7510
E-ISSN: 2581-690X
The arsenic (As) comprehensiveness in nature has aggravated the expansion of arsenic fortification and detoxification components in microorganisms. Many microorganisms discovered today with ability to oxidize arsenite (As3+) into arsenate (As5+) or reduce As5+ to As3+. In this study, two bacterial strains designated 3AB3 and 5AB2 was isolated from the soil samples collected from abandoned mining region of Madhya Pradesh and Jharkhand, India and arsenic concentration has been determined in both water and soil samples. Enrichment culturing method was employed for isolating bacteria and further they are screened for their redox ability. The isolated strains exhibited maximum growth at 30°C, at pH 7.0 in arsenic stressed Luria Bertani broth, checked through UV-Vis spectrophotometer at OD-620nm. Biochemical characterization of isolated strains was performed with various confirmation tests. Phylogenetic analysis of selected bacterial strains through MEGA-X confirmed their relationship to the genus Bacillus. Further, they are tested for transformation ability of arsenic (MSA method) and gene identification was done in selected isolated strains (PCR method). The result of this study shows that, even after abandoning the mining activities, concentration of arsenic increases in ground water by reducing ability of bacterial strains. PCR analysis depicted the presence of genes arsR, arsB and arsC in the strain 3AB3 and gene aoxB in 5AB2 respectively.
Arsenic, Mining, Gene, Transformation, Enrichment culture, Microplate Screening Assay (MSA), Bacillus
In recent years, improved industrial and agricultural activities have immensely enhanced a variety of chemicals in the environment. The contamination of ground water with arsenic (As) is a major concern in developing countries for the environment and to public health. Arsenic usually exist in underground region bounded with other minerals. Arsenite (As3+) is more mobile and toxic then arsenate (As5+)1-3. Its appearance in ground water is toxic above 10 µg/L (as said by WHO). If arsenite contaminated groundwater is consumed on regular basis, it gets accumulated in vital organs of the body disrupting their normal functions. Both plants and animals get severely affected by As3+ accumulation1,4,5. The mobility of As3+ is greatly affected by mining activities. So, for public health concern, government has abandoned mining activities in many regions. But even then also its concentration in underground water is increasing per year, as reported earlier. Since, ancient times, to overcome its toxic effects, bacterial species have developed specialized mechanism; the basis of which includes oxidation, reduction and methylation6,7.
Under anaerobic conditions, As5+ is reduced to As3+ via microbial activity and further gets methylated, forming monomethyl arsonic acid and then dimethylarsinic acid8,9. Solubility of arsenic is controlled by pH, biological activities, redox conditions and adsorption reaction10,11. The fresh water has arsenic ranges from 1-10 μg L-1, rising to 100 – 5000 μg L-1 in sulphur and iron mining area12,13.
At high redox abilities, arsenic becomes stable as As5+ oxyanions (H3AsO4 or H2AsO4–) but, under low redox potential and reducing conditions As3+ species (H3AsO3) becomes predominant14,15.
Native microbes utilizes As5+, electron acceptor during their respiration process. In the absence of oxygen, microbes gain energy by reducing As5+, resulting in production of toxic As3+ and expelling it out of cell in underground environment. Arsenate respiring organisms can affect water quality by catalyzing the mobilization of As3+, coupled with affecting biogeochemical cycles of other elements. So, our study aims to identify arsenic transforming bacterial species from abandoned mining regions of Madhya Pradesh (MP) and Jharkhand (JKD), India, with following objectives:
- To check the presence of arsenic in water and soil samples.
- To isolate and identify the arsenic oxidizing/reducing bacteria from contaminated sites.
- To characterize the arsenic transforming ability of bacterial isolates.
- To identify the genes responsible for arsenic transformation.
- Phylogenetic genetic relationship establishment using 16s rDNA gene sequencing.
The need for the remediation of Arsenic contaminated sites
Increased amount of arsenic was found on affected sites above acceptable limits. The World Health Organization (WHO 2001) has accepted limits of As in soil at 10 mg/kg and in water at 10 µg/L. This limit is far lower than what is realistic in numerous old mine locales and agricultural lands. In mining regions even when the mining activities are abandoned, then also the concentration of As increases.
The more mobile and toxic As3+ is converted to less mobile and less toxic As5+, by native bacterial species, in order to minimize the toxic As3+ concentration. The risk associated with mobile arsenic contaminants lies in its bioavailability through food chain in plants, microorganisms and consequently reaching to humans16-20. It is essential to decrease the arsenic level in soil, which can further decrease its ground water leaching and subsequent plant and human exposure. Many options were accessed for remediating contaminated sites such as landfill excavation, phytoremediation, solidification, and stabilization/immobilization, which were not proved to be significant and has their own limitations21. When the affected sites were visited, it was identified by talking to the local residents that, none of the person survived above 56 years of age. Even the crops grown on As contaminated soil also gets affected and when these plants are used as fodder for cattle, they gradually becomes weak and milk yielding capacity also decreases. In some animals As is also found present in milk22-25.
To deal effectively with arsenic, it is important to have a correct and proper understanding of factors and mechanisms controlling its accessibility in underground water and soil. This way, the problematic issues associated with arsenic can be efficiently controlled.
Regions Selected For Study
The different regions were selected based on the availability of high arsenic concentration either in soil or water. The selected regions are listed in Table S1 (supplementary file). The regions were selected based on the fact that even after abandoning the mining regions, arsenic concentration is increasing in contaminated regions26.
Sampling
A total of 112 samples (water + soil) were collected from contaminated abandoned mining regions of MP and JKD. Then, 250 ml and 400 gm of water and soil samples were collected aseptically in sterile capped bottles and transported to laboratory for analysis. The As content was found highest upto 3916 μg/Kg in soil and 1520 μg/l in water. On- spot qualitative analysis of arsenic was done using arsenic test kit (HiMedia Laboratories, India). Quantitative assessment of As was conducted by inductively coupled plasma – optical emission spectrometry (ICP-OES), technique through FICCI Research and Analysis Centre, Dwarka, New Delhi, India.
Table (1):
Arsenic concentration in collected water and soil samples – ICPOES (inductively coupled plasma atomic emission spectroscopy).
| S.no | Region | Place of Sample Collection | Source of sample | Water Sample (mg/L) | Soil Sample (mg/Kg) |
|---|---|---|---|---|---|
| 1 | Madhya Pradesh (MP) | Rajnandgaon | Water Supply Station | 1.0 | 12.40 |
| 2 | Abandoned Mining area Khairi | Borewell | 2.23 | 375.65 | |
| 3 | Abandoned mining area Kaudikasa | Borewell | 1.72 | 2.26 | |
| 4 | Jharkhand | Rajmahal | Drinking water Pond | 0.02 | 6.73 |
| 5 | Udhawa | Abandoned Mining area | 1.72 | 28.38 | |
| 6 | Dehari | Abandoned Mining area Tubewell | 1.2 | 31.84 | |
| 7 | Barhait | Abandoned Mining area Handpump | 0.05 | 0.51 |
Physico-chemical analysis of water and soil samples
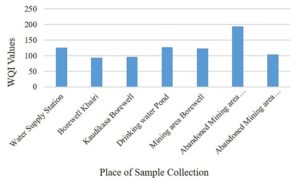
A total of 12 parameters were used to determine the physico-chemical properties of collected water samples (Table 2). These parameters were also used to determine water quality index (WQI), which is an important tool for determining quality of water for drinking purpose (Table S2 and S3, supplementary file). The graphical comparison between WQI of collected water samples from various regions is given in Fig. 1.
Table (2):
Summary of measured parameters in collected water samples.
Parameters |
Minimum |
Maximum |
|---|---|---|
pH |
5.2 |
8.4 |
Electrical conductivity (EC) |
615 |
1440 |
Total Dissolved Solute (TDS) |
438 |
1060 |
F– |
0.45 |
2.23 |
Cl– |
11.2 |
66.7 |
HCO3– |
180 |
565 |
SO42- |
28 |
490 |
NO3– |
0.6 |
42.3 |
Ca2+ |
26.4 |
130 |
Mg2+ |
17.6 |
194.7 |
Na+ |
14.8 |
188.4 |
K+ |
1.3 |
9.2 |
Total Hardness (TH) |
137 |
697 |
All concentration in mg/L, except pH and EC (µS/cm)
The water samples collected from the selected area is found acidic to alkaline in nature. The ionic balance of water is dominated by Mg2+, Ca2+, HCO3– and SO42-. In majority of the samples, the analyzed parameters are well within the desirable limits, but higher concentration of EC, TDS, SO42- and TH in many water samples make it unsafe for drinking purpose. The results suggested that the chemical composition of the water of study area is mainly controlled by environmental factors, weathering and with contributions from mining sources. The presence of arsenic tainted water at abandoned mining sites creates operational and stability problems, requiring effective water management strategy for drinking purpose.
Establishment of Enrichment Culture
Enrichment culturing technique was used to isolate microbial strains. Soil sample was inoculated in minimal salt medium (MSM) enhanced with 5 mM of sodium meta-arsenite (NaAsO2) after three successive serial dilutions. Basic components of MSM (mM l-1) were KH2PO4, 3.60; Na2HPO4.7H2O, 29.4; NaCl, 3.6; MgSO4, 1.66; FeSO4.7H2O, 0.0035; CaCl2, 0.009; H3BO3, 0.0098; NH4Cl, 18.69; CH3COONa, 60.95; Tryptone and Yeast extract (1.0 g l-1); Glucose (20.0 g l-1). The inoculated medium was incubated at 30°C and 120 rpm in rotary shaker. After seven days (168hr) of incubation, 5ml of culture was transferred to autoclaved MSM and incubated (30°C and 120 rpm) in rotary shaker. This process was repeated three times. The turbidity (denseness) of culture was analyzed by UV-Vis spectrophotometer, optical density at 620 nm (OD620) prior every sub-culturing. Later, cultures were consecutively diluted and inoculated on solid (agar containing) MSM for obtaining bacterial colonies.
Isolation of arsenic-resistant bacteria
After subsequent enrichment and growth of bacterial cells over solid MSM, they are then further grown on Luria agar (LA) media supplemented with 5mM of As3+ and As5+ respectively, followed by 96 hours of BOD incubation at 30°C. After 96 hours, white colored bacterial colonies were obtained. The physiological characteristics of bacterial colonies obtained are given in Table 3.
Table (3):
Physiological characteristics of bacterial colonies obtained.
| Physiological Characteristic |
Isolate 1 | Isolate 2 |
|---|---|---|
| 3AB3 | 5AB2 | |
| Shape | Circular | Circular |
| Elevation | Plane | Plane |
| Surface | Smooth | Smooth |
| Opacity | Opaque | Opaque |
Biochemical depiction
Morphological and functional characterization of isolated strains was executed through the standard approaches of Pelczar et al.27. Diverse biochemical properties of isolates like enzyme activity (indole, catalase, urease and oxidase), methyl red test, Voges–Proskauer test, citrate test, etc. were tested (Table 4), by following standard procedures27. Both the isolates were white in color, having flat colonies and smooth surface.
Table (4):
Biochemical Properties of the Bacterial Isolates.
| Parameters | Isolate 1 | Isolate 2 |
|---|---|---|
| 3AB3 | 5AB2 | |
| Catalase test | + | + |
| Indole test | – | – |
| Citrate test | – | – |
| Oxidase test | + | – |
| Urease test | – | – |
| Hydrogen sulphide production | – | – |
| Glucose fermentation | + | + |
| Arabinose test | + | + |
| Sucrose fermentation | + | + |
| Fructose | + | + |
| Inositol | + | – |
| Maltose | + | – |
| Mannose | + | + |
| Gram Staining | + | + |
Verification of Arsenic transformation – MSA Method
The ability of bacterial isolates to transform (reduce or oxidize) As3+ and As5+ respectively was determined by using microplate screening assay (MSA)28. Isolates were cultured on Luria agar (LA) plate containing, tryptone 1 g/l, beef extract 1 g/l, yeast extract 1 g/l and agar powder 15 g/l) with either As5+ or As3+ and incubated at 30°C for 96 hr. After incubation standard protocol was followed and small amount of silver nitrate (0.1M) was added to microplate. If the media turned brown, it confirms the presence of silver arsenate and if it turns yellow, the presence of silver arsenite was confirmed. Among the isolated strains 3AB3 and 5AB2 was confirmed found to reduce and oxidize arsenic respectively29.
16S rDNA sequencing for identification of bacterial isolate
PCR amplification of 16S rDNA (>1200 bp) and sequencing services were outsourced from National Centre for Microbial Research (NCMR), Pune. The obtained 16S rDNA sequence is submitted to Genbank database with accession number MH729060 of 3AB3 and MH729214 of 5AB2. Multiple Sequence Alignment and comparison with the 16S rDNA sequences available in GenBank database was performed using CLUSTAL W and BLAST programs, respectively. The phylogenetic tree was constructed using pairwise distance through MEGA X30.
Gene Identification
Gene identification was performed by PCR method which was carried out in Allele Life Sciences, Noida, India and the respective results were obtained.
Table (5):
Details of target gene, primer used and amplicon size.
Gene target |
Function |
Primer F & R |
%GC |
Melting Temper-ature (0C) |
Amplicon size (bp) |
Marker (bp) |
|---|---|---|---|---|---|---|
aoxB |
Arsenite oxidase |
F – TACGATCGGCTGCAATTCCC R – GCACCATTGGGAACGATGTG |
60 |
60 |
395 |
100 |
arsR |
Ars operon regulator |
F – TTAATGGAAAAACAATTGAAGGCTG R – GCCCTCCACACATTCAGAGTT |
50 |
57 |
300 |
100 |
arsB |
Membrane associated efflux pump |
F – TTGTGTTAGCGATGGTGCGT R – ATCCTGCATTGCCAAGTCCG |
60 |
60 |
540 |
100 |
arsC |
Arsenate reductase |
F – CCGGAAGACTGGGAAGTGT R – GCTGCATCCCCACAAAGTGT |
57 |
58 |
170 |
100 |
The presence of arsenic in water and soil samples was determined using inductively coupled plasma atomic emission spectroscopy (ICPOES) technique (Ficci Frac Laboratory, New Delhi) which was ranged between 1000 µgL-1 to 1520 µgL-1 in water and 2000 μgKg -1 to 3916 μgKg -1 in soil samples. All the samples collected was found alkaline to basic property with an average of pH 6.8. The bacterial cells were isolated through enrichment culturing technique using MSM media. The bacterial colonies (Fig. S1, supplementary file) were obtained on LA plates, standardized to pH 7.2.
Physico – chemical Properties of water and soil
Drinking water of good quality is the basic requirement for human physiology. Even the government has also started the provision of mobile water supply to rural as well as urban population to meet its necessity and to prevent health problems. Groundwater is generally considered fresh and safe source of drinking water, but rapid population growth and increased industrialization have resulted in greater contamination of quality water.
A total of 112 samples (water and soil) were collected from abandoned mining regions of MP and JKD in sterile bottles. The physiological properties of both water and soil is determined upon pH, temperature, TDS etc. (Table 2). The most important factor i.e. As concentration in the abandoned mining regions has also been determined commercially by ICPOES technique. The concentration of As in both water and soil samples is given in Table 1.
Isolation of Arsenic resistant bacterial isolates
Enrichment cultures were established in 500 ml Erlenmeyer flask containing 100 ml of autoclaved MSM, supplemented with 5 mM of As3+ and As5+ salt, separately. The flask were then kept at 120 rpm on a rotatory incubator shaker at 30°C. The enrichments were sub-cultured weekly into a freshly prepared MSM containing same concentration of arsenate and arsenite. After the second sub-culture, the enriched bacterial cultures were serially diluted and plated onto L.A medium (1.5%) containing 5 mM of As3+ and As5+. Single colonies were picked and streaked onto same medium to obtain pure colonies Fig. S1 (supplementary file).
Transformation of arsenic (MSA method)
Tris-based buffer was used to standardize the micro-plate assay, irrespective of growth medium, to prevent interference problems with other ions, present in growth (MSM) media, which could result in color formation with silver nitrate (AgNO3) under the same conditions similar to arsenic ions.
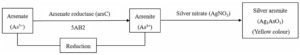
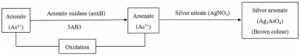
After adding AgNO3 in required wells of microtitre plate, it was incubated for 96 hr. The 3AB3 culture plate media having arsenite, gradually turned brown (Fig. 2) confirming the existence of silver arsenate formed by conversion of As3+ to As5+ in the media. At the point when silver nitrate was mixed with arsenate containing culture, it turned yellow confirming formation of silver arsenite by conversion of As5+ to As3+.
Well 1 (row – 4): inoculated with bacterial isolate 1 (designated as 5AB2), Well 2 (row- 4): blank without bacterial cells, Well 2 & 3 (row – 5): blank without bacterial cells, Well 4 (row – 4): blank without bacterial cells, Well 4 (row – 5): inoculated with bacterial isolate 2 (designated as 3AB3).
Some other isolates were also obtained which didn’t show any type of transformation. Among two isolated bacterial strains, the strain present in Well number 1 (row- 4) has given positive result and designated as 3AB3 and in Well number 4 (row – 5) designated as 5AB2 (Fig. 2). Hence, it was confirmed that bacteria named 5AB2 possess the skill to reduce As5+ to As3+ and bacteria named 3AB3 oxidized As3+ to As5+. A clear representation of mechanisms intended by bacterial isolates for arsenic transformation are given in fig. 3.
Fig. 3(a). Mechanism of arsenate to arsenite transformation by 5AB2 bacterial isolate
Fig. 3(b). Mechanism of arsenite to arsenate transformation by 3AB3 bacterial isolate
Fig. 3. Mechanism of arsenic species transformation by 5AB2 (3a) and 3AB3 (3b) bacterial isolates. This transformation mechanism was studied using microplate screening assay
Phylogenetic analysis
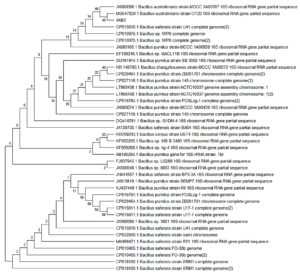
The phylogenetic history of 3AB3 and 5AB2 named bacterial isolates was retrieved (Fig. 4 and 5) using Neighbor-Joining method31. The phylogenetic tree was constructed using 500 duplicates representing evolutionary past32. The evolutionary distances was intended using Maximum Composite Likelihood method using MEGA X software33,34. The phylogenetic analysis revealed that the two isolated strains 3AB3 and 5AB2 are closely related to Bacillus sp.
Fig. 4. Phylogenetic tree of strain 3AB3 together with its related neighbors was constructed based on 16S rDNA sequence using neighbor-joining and maximum likelihood method. GenBank accession numbers are given in parentheses.
Fig. 5. Phylogenetic tree of strain 5AB2 together with its related neighbors was constructed based on 16S rDNA sequence using neighbor-joining and maximum likelihood method. Genbank accession numbers are given in parentheses.
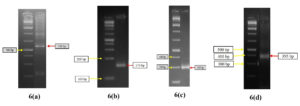
Fig. 6. Gel doc images of ars operon components. arsB (6a), arsC (6b) and arsR (6c) forms main component of ars operon. Fig 6(d) is gel doc image of aox operon component aoxB. The aox operon has two components aoxA and aoxB, aoxB is responsible for transformation of arsenite to arsenate3.
Gene Identification
PCR method was used with specified primers, designed by Primer blast tool of NCBI and the experiment was performed commercially. For conduction of PCR, the primer sequences and respective bacterial samples were brought to Allele life sciences, Noida and following (Fig. 6) results were obtained.
- The genes of 5AB2 (transforms As5+ to As3+) was identified to be arsB, arsC, arsR (Fig. 6a, 6b, 6c)
- The genes of 3AB3 (transforms As3+ to As5+) was identified to be aoxB (Fig. 6d)
The present work focusses on the identification and transformation potential of arsenic sp. (As3+ and As5+). To accomplish the work, water and soil samples were collected from arsenic contaminated abandoned mining regions of MP and JKD. The collected samples (water + soil) were analyzed for the presence and absence of arsenic through ICPOES (FICCI Research and Analysis Centre, New Delhi). After analysis the samples found positive for containing arsenic were processed and bacterial strains were isolated through enrichment culturing method using minimal salt medium (MSM). Isolated bacterial strains were biochemically characterized (Table 4) and were named as 3AB3 and 5AB2 respectively. The individual strains performing transformation, As3+ to As5+ and As5+ to As3+ was identified using Tris-HCl through MSA method. The percent transformation has also been determined in fig. S2 (Supplementary file), which depicts the time taken to transform individual arsenic forms by isolated bacterial strains. The 16s rDNA sequencing of selected transformants were performed at National Centre for Microbial Resource, Pune, India, and the result was obtained accordingly. The obtained 16s rDNA sequence of respective bacterial isolates are submitted to Genbank with accession numbers MH729060 (3AB3) and MH729214 (5AB2) respectively. The phylogenetic relationship was also established using CLUSTAL W through MEGA-X software (Fig. 4 and Fig. 5). The isolated bacterial strains shows the closest relationship with the Bacillus sp. The genetic identification through PCR method revealed that isolate 3AB3 contains the gene aoxB, which is responsible for conversion of As3+ to As5+. This oxidizing operon is not well studied and needs further study, as it contains many other intermediate sequences for which the function is not known. Another isolated bacterial strain 5AB2, which is reported here for converting As5+ to As3+, harbors the main components of ars operon viz, arsR, arsB and arsC. The details of all these genes are provided in Table 5.
The mobility and toxicity of As3+ is far more than As5+ and it is also considered to be lethal for plant and human on consumption35-37. The bacterial strain 3AB3 was identified for transforming toxic As3+ to As5+ and 5AB2 was identified to be converting As5+ to As3+, which shows that isolated bacterial cells possess the mechanism to encounter arsenic toxicity and to overcome it. This transformation mechanism of bacterial cells needs further study and can also be utilized for biological remediation of toxic arsenic species.
Thus, this complete study, concludes that two microbial isolates 3AB3 and 5AB2 were identified nearest to Bacillus australimaris (NH7I-1) and Bacillus subtilis sub sp. Stercoris (D7XPN1) have 97.09% and 99.92% similarity. The isolated, biochemically characterized and identified bacterial isolates were recognized for their transformation ability. This result gives us the idea that even after quieting the mining work from the specified sites, the concentration of As3+ increases by bacterial transformation mechanism. The MSA test resulted oxidizing ability of isolate 3AB3 transforming harmful As3+ to As5+. The As5+ has already been recognized having less toxicity than arsenite on plants and human health, if consumed. So, this test also gives us the idea of effective elimination of toxic As3+, improving soil and water quality. The phylogenetic relationship was established, which depicts that 3AB3 and 5AB2 are closely related to Bacillus sp. containing oxidizing and reducing property. The potential bacterial mechanism of encountering As toxicity by altering its oxidation state and removing its noxious effects and acquiring resistant characteristic is an exceptional property of bacterial cells and can be utilized in the innovative strategy for bioremediation of this heavy metal.
Additional file: Additional Table S1- S3. Additional Figs. S1 and S2.
ACKNOWLEDGMENTS
We would like to express our heartfelt thanks to Er. Ankit Singh for providing assistance on methods. Thanks to FICCI Research and Analysis Centre, New Delhi for providing assistance on quantitative analysis. We would like to thank the Department of Biotechnology and Life Sciences, Mangalayatan University and technical staff for extending the support for the completion of this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Upadhyay MK, Shukla A, Yadav P, Srivastava S. A review of arsenic in crops, vegetables, animals and food products. Food chem. 2019;276:608-618.
Crossref - Achour AR, Bauda P, Billard P. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res in Microbiol. 2007;158(2):128-137.
Crossref - Cai L, Rensing C, Li X, Wang G. Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers: Achromobacter sp. SY8 and Pseudomonas sp. TS44. Appl Microbiol Biotechnol. 2009;83(4):715-725.
Crossref - Ravenscroft P, Brammer H, Richards K. Arsenic Pollution: A Global Synthesis. 2009;1.
https://doi.org/10.1002/9781444308785 - Chen YW, Yu X, Belzile N. Arsenic speciation in surface waters and lake sediments in an abandoned mine site and field observations of arsenic eco-toxicity. J Geochem Explor. 2019;205:106349.
Crossref - Upadhyay MK, Yadav P, Shukla A, Srivastava S. Utilizing the potential of microorganisms for managing arsenic contamination: A feasible and sustainable approach. Front Environ Sci. 2018;6:24.
Crossref - Costa M. Review of arsenic toxicity, speciation and polyadenylation of canonical histones. Toxicol Appl Pharmacol. 2019;375:1-4.
Crossref - Pongratz R. Arsenic speciation in environmental samples of contaminated soil. Sci Tot Environ. 1998;224(1-3):133-141.
Crossref - Siddiqui SI, Chaudhry SA. A review on graphene oxide and its composites preparation and their use for the removal of As3+ and As5+ from water under the effect of various parameters: Application of isotherm, kinetic and thermodynamics. Proc Safe Environ Protect. 2018;119:138-163.
Crossref - Xie Z, Wang J, Wei X, et al. Interactions between arsenic adsorption/desorption and indigenous bacterial activity in shallow high arsenic aquifer sediments from the Jianghan Plain, Central China. Sci Tot Environ. 2018;644:382-388.
Crossref - Smedley PL, Kinniburgh DG. A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem. 2002;17(5):517-568.
Crossref - Chakraborti D, Biswas B, Chowdhury TR, et al. Arsenic groundwater contamination and sufferings of people in Rajnandgaon district, Madhya Pradesh, India. Curr Sci. 1999;77(4):502-504.
- Mandal BK, Suzuki KT. Arsenic round the world: a review. Talenta. 2002;58(1):201-235.
Crossref - Saxena VK, Kumar S, Singh VS. Occurrence, behaviour and speciation of arsenic in groundwater. Curr Sci. 2004:281-284.
- Li Y, Li W, Xu H. Speciation, Activity, Transformation, and Degradation of Heavy Metals and Organochlorines in Red Soils. In: Luo Y, Tu C (eds) Twenty Years of Research and Development on Soil Pollution and Remediation in China. Springer Singapore. 2018;735-762.
Crossref - Anawar HM, Garcia-Sanchez A, Murciego A, Buyolo T. Exposure and bioavailability of arsenic in contaminated soils from the La Parrilla mine, Spain. Environ Geol. 2006;50(2):170-179.
Crossref - Naidu R, Smith E, Owens G, Bhattacharya P. Managing arsenic in the environment: from soil to human health. CSIRO publishing. 2006.
Crossref - Ghosh G, Mukhopadhyay DK. Human Health Hazards Due to Arsenic and Fluoride Contamination in Drinking Water and Food Chain In: Sikdar P (eds) Groundwater Development and Management. Springer Cham. 2019; 315-369.
Crossref - Sanyal SK. Arsenic Contaminated Irrigation Water and Its Impact on Food Chain. In: Sikdar P (eds) Groundwater Development and Management. Springer Cham. 2019;309-327.
- Moulick D, Santra SC, Ghosh D. Effect of selenium induced seed priming on arsenic accumulation in rice plant and subsequent transmission in human food chain. Ecotoxicol and Environ Safe. 2018;152:67-77.
Crossref - Liu L, Li W, Song W, Guo M. Remediation techniques for heavy metal-contaminated soils: principles and applicability. Sci Tot Environ. 2018;633:206-219.
Crossref - Raessler M. The arsenic contamination of drinking and groundwaters in Bangladesh: featuring biogeochemical aspects and implications on public health. Arch Environ Contaminat Toxicol. 2018;75(1):1-7.
Crossref - Zhou X, Zheng N, Su C, Wang J, Soyeurt H. Relationships between Pb, As, Cr, and Cd in individual cows’ milk and milk composition and heavy metal contents in water, silage, and soil. Environ Pollut. 2019;255:113322.
Crossref - Bassil M, Daou F, Hassan H, et al. Lead, cadmium and arsenic in human milk and their socio-demographic and lifestyle determinants in Lebanon. Chemosphere. 2018;191:911-921.
Crossref - Kambli A, Sole S, Garud K, Lonare S, Bagal N. Quantitative analysis of toxic metals in buffalo milk samples from mumbai suburban region by ICP_AES. J Anim. Health Prod. 2019;7(1):5-10.
Crossref - Hajalilou B, Mosaferi M, Khaleghi F, Jadidi S, Vosugh B, Fatehifar E. Effects of Abandoned Arsenic Mine on Water Resources Pollution in North West of Iran. Health Promotion Perspect. 2011;1(1):69-72.
- Riker AJ, Warren J, Weeks OB. Manual of microbiological methods. Society of American Bacteriology. McGraw Hill Book Company Inc. New York. 1957. https://archive.org/stream/manualofmicrobio00amer/manualofmicrobio00amer_djvu.txt
- Simeonova DD, Lievremont D, Lagarde F, Muller DA, Groudeva VI, Lett MC. Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol Lett. 2004;237(2):249-253.
Crossref - Dey U, Das K, Roy P, Chatterjee SN, Mondal NK. Searching of microbial agent for bioremediation of arsenic. Int J Ext Res. 2015;5:60-64.
- Kumar S, Stecher G, Tamura K. MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceed Nat Acad Sci. 2004;101(30):11030-11035.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Shakoor MB, Riaz M, Niazi NK, et al. Recent Advances in Arsenic Accumulation in Rice. In (edn) Advances in Rice Research for Abiotic Stress Tolerance Elsevier. 2019; 385-398.
Crossref - de Souza ACM, de Almeida MG, Pestana IA, de Souza CMM. Arsenic Exposure and Effects in Humans: A Mini-Review in Brazil. Arch Environ Contaminat Toxicol. 2019;76(3):357-365.
Crossref - Alexander J and Oskarsson A. Toxic metals Chemical hazards in foods of animal origin. Wageningen Academic Publishers. 2019; 67-73.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.