Snake venom has developed over millions of years as a tool for capturing prey and defending against predators and other threats. Snake venom contains diverse proteins and peptides, which elicit a range of pathophysiological abnormalities including cytotoxic, neurotoxic, myotoxic, hemolytic and inflammatory effects in the victim, and cause severe morbidity. Although these venom proteins have significant pharmacological potential, many remain insufficiently explored for therapeutic applications. Therefore, this article critically reviews the attributes of selected toxic proteins, which make them suitable drugs for the well-being of mankind. Although these toxins have serious adverse effects on human health, research has shown that they can be modified to exhibit beneficial properties and serve as potential treatments for various diseases. In addition to being a model protein for drug development, the similarity in sequence and structure indicates that these toxic proteins can be used in protein replacement therapy to combat several human diseases. The article also addresses the challenges faced during the entire process, starting from the initial phase of choosing venom proteins to drug formulation. While nanotechnology-based formulations of snake venom-derived drugs exhibit promise across diverse therapeutic domains, additional research and development efforts on the different variants of these proteins are essential to fully unlock their clinical advantages and enhance their efficacy for disease treatment.
Snake Venom Protein, Therapeutic Potential, Human Disease, Protein Similarity
In 2017, the World Health Organisation officially classified snakebite as a neglected tropical disease (NTD). During a snakebite, venom – a specialised toxic secretion is injected into the victim. Globally, snakebites affect up to 2.7 million people annually, leading to estimated 81,000 to 138,000 deaths yearly.1 Survivors of snakebites often face long-term consequences, with the burden of premature death and disability measured at 6.07 Disability-Adjusted Life Years (DALYS).2 Prevention and treatment of snakebites can be achieved through education, improved access to antivenom, and timely medical intervention by trained healthcare workers, effective governance, and related measures. Conventional surveillance systems reveal that India records the highest number of snakebite-related fatalities, accounting for half of all global cases and contributing to 2.97 million DALYs. Around 70% of these fatalities occur in eight states – Bihar, Jharkhand, Madhya Pradesh, Odisha, Uttar Pradesh, Andhra Pradesh (including Telangana), Rajasthan, and Gujarat, which together account for more than half of India’s population. Among these, Odisha is a critical focus of India’s Avoidable Death Network (ADN) Hub, as snakebite remains a significant and pressing public health concern in the state.3
The interest in PLA2, LAAO, hyaluronidase, and metalloproteinase is driven by their critical contributions to the lethality of snake venom. Their mechanisms of action reveal how venom can rapidly immobilize or kill prey while also offering insights into physiological processes that can be harnessed for therapeutic purposes. Their study provides opportunities for improving antivenoms and exploring novel medical applications based on their lethal properties. The major venom proteins present in different snake toxin families are PLA2‚ SVMP, SVSP, and three-finger proteins.4-6 SVSPs cause a wide range of effects, including coagulation activation, inflammation, and tissue damage. Some SVSPs can interact with other venom proteins to enhance their activity. Other proteins in the snake venom include LAAOs, Kunitz peptides, disintegrins, NP, cysteine-rich secretory proteins, and C-type lectins.
Some of the snake venom proteins have potential for drug development but face challenges in being toxic and storage instability. Some venom proteins resemble human proteins, offering targeted treatment possibilities. Lack of detailed structure-function studies limits therapeutic use and large-scale production. Current antivenoms are broad-spectrum and often miss specific toxins. Integrating venomics data can help create more precise and effective antivenoms. Research centered on exploring the chemistry and understanding the relationship between protein structure and function. Proteins and peptides found in snake venom, as well as the exploration of their potential therapeutic applications, have gained considerable attention in biomedical research.7 Recent venom-based drugs currently on the market or in clinical trials have been reviewed.6,8 Though these review articles provide valuable information, the significance of snake venom proteins as therapeutic agents can be better exploited if the sequence and structure correlations of these proteins, especially in relation to the human proteins, are completely understood. This review examines the similarities in amino acid sequences and structures between major snake venom proteins and human proteins. Further, challenges encountered during the development of drugs from venom proteins and the therapeutic potentials of the various venom proteins for the treatment of different diseases are discussed. To provide a holistic view, we have also included some basic information about the venom proteins, especially the venom composition and the chemistry of selected enzymatic toxins, which will benefit the readers. This study will provide insight into the use of enzymatic toxins as a substitute for non-functional human proteins occurring during various disease states.
The complex chemistry of Snake venom
Although there are notable exceptions and considerable species-level variability, certain toxic proteins play a key role in the effects of snake venom following a bite. 3FTx and PLA2‚ are the major components of elapid venoms, and comprise about 65%-70% of the total proteins of the venom proteome. Whereas, snakes from the Dendroaspis genus (mambas) and various Australian snakes exhibit significant deviations in the composition and function of these toxins. Mambas’ venoms lack PLA2, while venoms of Australian snakes have a notably low content (<6%) of 3FTx. On average, 6% of elapid venom is made up of SVMPs,9,10 snake venom serine proteases (SVSPs),11,12 and L-amino acid oxidases (LAAOs).13-15 About 5% of elapid venom is made of Kunitz-type peptides,12,16 a group of serine protease inhibitors recognized by the presence of the Kunitz domain fold that are specific K+-channel blockers. Mambas are extremely rich in Kunitz-type peptides. Approximately 10%-12% of the proteins are mostly non-toxic proteins, with highly variable composition11,17-19 and the reason for the occurrence of many of these proteins in snake venom is not fully known.
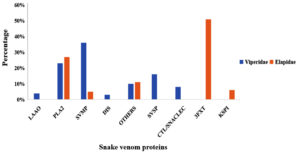
The venoms of viperid snakes encompass toxins from nine distinct protein families, such as PLA2‚ SVMPs, SVSPs, C-type lectins (CTLs), LAAOs, Disintegrins, Three-Finger Toxins, Kunitz-type peptides, CRISPs families.20-22 Again, there are numerous exceptions and considerable variability in toxin content in venoms within each species and subspecies of snakes. PLA2‚10,11 SVMP,9-11,19 and SVSP11,23 toxins are prevalent in the majority of species, accounting for an average of 70% of the total venom proteome. Although viperid PLA2‚ share higher sequence homology with the neurotoxic PLA2‚ found in elapids, the majority of viperid PLA2‚ exhibit myotoxic effects. LAAOs,13-15 C-type lectins and C-type lectin-like proteins,24 and natriuretic peptides,6,17 are other toxins present in smaller quantities (4%-7%) (Figure 1), in the Viperid family. Reports indicate that snake venom toxins act synergistically. The pathophysiology of snakebite envenomation is determined by the combination and relative proportions of various toxins present in the venom.2,25 Hemotoxicity, myotoxicity, cardiotoxicity, neurotoxicity, and cytotoxicity are the results of snakebite envenomation. Further, these four major toxic proteins phospholipase A2‚ (PLA2), snake venom metalloproteases (SVMPS), snake venom hyaluronidase and L-amino acid oxidase (LAAO) from snake venom have been reported to have both physiological effects and therapeutic potential in humans. The intricate interplay between the components of snake venom and their varied concentrations among species underpins their diverse pathophysiological effects. The detailed chemistry of venom proteins not only elucidates the mechanisms of envenomation but also highlights their promise as a source of novel drugs. Combining insights from venom composition with molecular studies of individual toxins bridges the gap between understanding venom pathology and harnessing its benefits for human health.
Figure 1. Graphical representation showing the distribution of venom components in two snake families: Elapids and Viperids. The X-axis displays different venom protein types, while the Y-axis indicates their percentage composition in snake venom. Data, primarily highlighting SVMP, PLA2, and SVSP as dominant components, are sourced from the Isbister and Tasoulis database and literature; non-toxic families were excluded
Key Toxic Proteins in Snake Venom: Chemistry and Mechanisms
Snake venom phospholipase
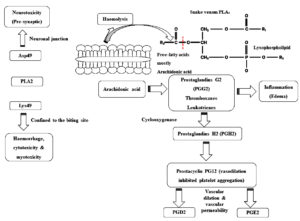
Phospholipase A (PLA2) enzymes cause a range of pathological effects. PLA2‚ exerts distinct effects on the nervous and cardiovascular systems through high-affinity interactions with specific receptors, a mechanism that is independent of its enzymatic function. Once attached to the target, they produce pharmacological effects with or without catalytic activity.26 PLA2‚ in snake venom cause neurotoxicity, thus damaging the neurons at the presynaptic cleft. Bulk of pre-synaptic neurotoxins derived from snake venom discovered so far are protein complexes containing PLA2. When they bind to receptors or lipid domains in the motor neuron plasma membrane at the neuromuscular junction, they induce changes in membrane activity. This modification in membrane potential facilitates the influx of Ca2+ from the extracellular environment. The resulting changes in membrane permeability enhance the exocytosis of synaptic vesicles (Figure 2).27
Figure 2. Mechanism of action of snake venom PLA2. Aspartate (Asp) at position 49 (D-Asp49 type PLA2) and Lysine (Lys) at position 49 (K-Lys 49 type PLA2), Platelet-Activating Factor (PAF): platelet
Myotoxic svPLA2‚ generally possess aspartic acid at position 49, which is important for their enzymatic activity. Asp49-PLA2‚ are catalytically active, but Lys49 homologs exhibit either weak catalytic activity or are catalytically inactive. Additionally, substitutions such as Ser49, Arg49, Asn49, or Gln4928 have been identified and are known to alter catalytic activity, thereby influencing the toxic effects of this venom protein. Asp49 PLA2‚ hydrolyzes phospholipids to release lysophospholipids, which leads to the skeletal muscle necrosis by directly disrupting membrane stabilization and/or indirectly altering membrane biophysics.29 On the other hand, Lys49 PLA2‚ myotoxins do not have catalytic activity and exist as homodimers.30 The C-terminal regions (residues 115-129) of these peptides are rich in basic, aromatic, and hydrophobic amino acids, which are responsible for their myotoxic effects.31,32 Site-directed mutagenesis studies have identified Tyr117, Arg118, Tyr119, Lys122, and Phe125 as key residues significantly influencing myotoxicity.33
Venom protein PLA2‚ variants can broadly be divided into D49 acidic PLA2‚ (Asp-49), S49 PLA2‚ (Ser-49), and K49 basic PLA2‚ (Lys-49 replacing Asp-49). Basic PLA2‚ homologs, whose amino acid residues, such as K49 and S49 PLA2‚ are involved in different Ca²+ independent functions, but they are still catalytically inactive.34 Among these, K49 basic PLA2‚ exhibits higher cytotoxicity compared to D49 acidic PLA2. Generally, acidic PLA2‚ has a higher IC50 value than basic PLA2, indicating lower cytotoxic potency.
S49 PLA2‚ variants, identified in the venom of saw-scaled vipers (Echis sp.), exhibit enzyme activity that is independent of calcium ions and exhibits higher toxicity to cells in comparison to K49 PLA2‚ (IC50 = 2.5-12.2 µM).35 However, K49 PLA2‚ demonstrates greater lipolytic activity compared to S49 PLA2.35
Snake venom metalloproteases (SVMPs)
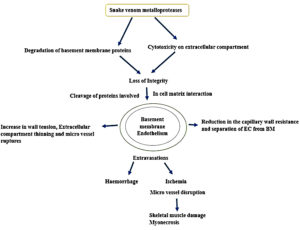
SVMPs account for 30%-60% of the snake venom in the Viperidae family and are categorized into three distinct classes (P-I, P-II and P-III) mainly depending on their organization of the protein domain and therefore their functional variation. All classes of SVMPs contain the zinc ion-binding domain, which is involved in enzymatic activity. P-I SVMPs exclusively possess the zinc ion-binding domain and exhibit molecular weight range of 20 to 30 kDa. However, P-II and P-III SVMPs contain an additional domain that confers additional functional characteristics to these proteins. P-II SVMPs contain a disintegrin-like domain and have a molecular weight of 30-60 kDa, whereas P-III SVMPs, have a molecular weight in the range of 50 to 70 kDa, are composed of a cysteine-rich and disintegrin-like domain. Some of them have a quaternary structure, whereas a disulfide bond of a smaller lectin type C subunit is linked to the P-III SVMP subunit.36 These snake venom metalloproteases activate prothrombin, defibrinogenesis, and coagulation X factor, which results in fibrinolytic stimulation.37,38 They contribute actively to the breakdown of the extracellular matrix and induce an inflammatory response.39 Snake venom metalloproteases have the potential to act as thrombolytic agents. For example, a fibrinolytic enzyme isolated from Agkistrodon contortrix has potent fibrinolytic activity (Figure 3). Alfimeprase recombinant form is employed in the treatment of abnormal blood clot formation by efficiently promoting thrombolysis in case of acute limb ischemia.40
Snake venom hyaluronidase
Hyaluronidases are a diverse group of enzymes found in various organisms, primarily responsible for cleaving hyaluronic acid, a glycosaminoglycan molecule of high molecular weight. Some of these enzymes can also degrade other glycosaminoglycans, although at a slower rate.41 In snake venom, hyaluronidases function as spreading factors during envenomation, when hyaluronic acid present in the extracellular matrix of the local tissues gets broken down into smaller molecules. These venom hyaluronidases are classified as neutral endo-β-N-acetyl-D-hexosaminidases, with a different range of molecular weight from 33 kDa to 110 kDa. They display structural diversity due to post-translational modifications or truncation of transcripts. The translated proteins typically consist of 440 to 450 amino acids.42 Snake venom hyaluronidase facilitates the breakdown of internal glycoside bonds in acidic muco-polysaccharides and decreases the integrity and viscosity of connective tissues. As the hyaluronic acid barrier is ruptured, other venom components penetrate the tissues effectively. The breakdown of HA in the extracellular matrix enhances membrane permeability, making tissues more susceptible to injected fluids.43
L-amino acid oxidase (LAAO)
The homodimeric flavoenzyme LAAO (EC 1.4.3.2) contains covalently linked flavin adenine dinucleotide and renders the snake venom its characteristic yellow colour (FADs). Each LAAO subunit has a molecular weight of 50-70 kDa and 110-159 kDa in its dimeric state.44,45 The three major domains of the LAAO are substrate binding, FAD binding, and helical domains. It has been reported that the substrate-binding domain contains 7 mixed β-pleated sheets.46 The enzyme engages with amino acid residues (serving as substrates) and oxygen to create a ternary complex, subsequently catalyzing the reduction of flavin semiquinone. During the reductive half-reaction, the FAD cofactor undergoes re-oxidation, generating hydrogen peroxide in the process, as described by Kang.18
The structural and functional resemblance of snake venom proteins to human counterparts allows them to interact with human physiological systems with high specificity. These similarities explain their potent effects in envenomation and their emerging therapeutic applications. Snake venom proteins exploit conserved biochemical pathways in humans, such as membrane dynamics (PLA2), matrix remodelling (SVMPs), redox processes (LAAOs), and tissue permeability (hyaluronidases). Understanding the chemistry of venom proteins and their mimicry of human proteins aids in repurposing them for medical use, such as anti-inflammatory agents (PLA2), thrombolytics (SVMPs), cancer therapeutics (LAAOs), and drug delivery systems (hyaluronidases). Snake venom proteins often exhibit enhanced efficiency and potency compared to their human analogues, providing a blueprint for designing synthetic or recombinant therapeutics. By linking the chemistry of these toxins to their similarities with human proteins, researchers can harness venom proteins’ dual roles-mitigating their toxic effects while leveraging their therapeutic potential.
Similarities between snake venom toxins and human proteins
As indicated earlier, understanding the sequence and structural similarities of the selected snake venoms and the human counterparts will be of high value for exploiting the therapeutic benefits of snake venom proteins. Investigating the similarity of human proteins with snake venom proteins will be of potential help to design the model compounds for targeting human diseases and to develop enzyme-based replacement therapy for the enzyme-deficient disorder or diseases in humans.
Phospholipase A2 (PLA2)
Sequence analysis reveals that human pancreatic phospholipase A2‚ (accession number 3ELO_A) has the highest sequence similarity with svPLA2‚ of Naja naja species (UniProt accession number P15445) with 98% query coverage, 48.03% identity and 69.4% similarity (Figure 4). However, the lowest sequence similarity is with group IIF secretory phospholipase A‚ isoform X1 (Homo sapiens) (accession number XP_011540257.1) with 63% query coverage, 38.67% identity and 48.0% similarity. However, snake venom PLA2‚ (svPLA2) enzymes share significant structural and functional similarities with human PLA2‚ such as high catalytic activity, specificity towards phospholipids, and pro-inflammatory responses.47 The domain structure of snake venom PLA2‚ varies between different species, but generally consists of a calcium-binding loop, a helical domain, and a catalytic domain. The calcium-binding loop stabilizes the reactive intermediate,48 while the helical domain stabilizes the enzyme structure and contains residues involved in substrate recognition. The amino acids in the catalytic domain are highly conserved and are responsible for phospholipid hydrolysis, and are highly specific to phospholipids with fatty acid chains of 16-20 carbon atoms.26 In comparison, human PLA2‚ also consists of a calcium-binding loop, a helical domain, and a catalytic domain, but has greater structural complexity and diversity due to alternative splicing, post-translational modifications, and the presence of multiple isoforms with different tissue expression patterns and substrate specificities. Human PLA2‚ is classified into several subfamilies, including cytosolic PLA2 (cPLA2), calcium-independent PLA2 (iPLA2), secreted PLA2‚ (sPLA2), and platelet-activating factor acetyl-hydrolase (PAF-AH).49 There are differences in the amino acid sequence and 3D structure that confer distinct biochemical and pharmacological properties to these enzymes.50 For example, some svPLA2‚ enzymes have a myotoxic or neurotoxic effect, while human PLA2‚ is involved in the regulation of various physiological mechanisms such as inflammation, lipid metabolism, and cell signalling. With the identification of diverse targeted delivery systems with high specificity, the enzyme replacement therapeutic approaches are highly feasible to combat the pathophysiological effects related to the underexpression of PLA2.
Figure 4. Sequence alignment for the snake venom PLA2 and the corresponding human protein, which has the most similarity. In the figure, the protein with UniProt ID P15445 represents a snake venom Naja naja species PLA2, while 6Q42 corresponds to a human PLA2 protein
Metalloproteinase
Sequence analysis indicates that ADAM metallopeptidase domain 28 (Homo sapiens) (accession number XP_011542671.1) has the highest sequence similarity with snake venom metalloprotease of Naja atra species (UniProt accession number D3TTC2) with 96% query coverage, 43.16% identity and 58.5% similarity (Figure 5). Lowest sequence similarity is with ADAM metallopeptidase domain 9 (meltrin gamma), the isoform CRA_a (H. sapiens) (accession number EAW63283.1) with 68% query coverage, 36.74% identity and 54.0% similarity. Snake venom metalloproteases (SVMPs) and human metalloproteases (HMPs) are part of the broader metalloproteinase family, characterized by the presence of a zinc ion in the enzyme’s active site. In Class P-I SVMPs, alongside the catalytic domain, there is a disintegrin domain responsible for inhibiting platelet aggregation. Class P-II SVMPs feature a cysteine-rich domain that facilitates binding to the extracellular matrix, while Class P-III SVMPs encompass both a disintegrin-like domain and a cysteine-rich domain. In contrast, human HMPs are divided into six groups based on their domain structure. These metalloproteases play crucial roles in the regulation of various physiological mechanisms such as tissue regeneration and wound healing. However, their dysregulation has been implicated in various pathological conditions, including cancer and inflammatory diseases. Similarly, SVMPs contribute to the pathogenesis of oedema, inflammation, myonecrosis, skin damage, and the onset of cardiovascular failure.22,51,52
Figure 5. Sequence alignment for the snake venom metalloprotease and the corresponding human protein, which has the most similarity. In the figure, the protein with UniProt ID D3TTC2 represents a snake venom metalloprotease from Naja atra species, while XP_011542671.1 corresponds to a human metalloprotease protein
Hyaluronidase
Sequence analysis indicates that hyaluronidase 3 and hyalurono-glucosaminidase 3 of Homo sapiens (accession number KAI2529754.1) has the highest sequence similarity with snake venom hyaluronidase of Ophiophagus hannah species (UniProt accession number V8PG63) with 82% query coverage, 48.65% identity and 63.0% similarity (Figure 6). Lowest sequence similarity is with hyaluronidase 2, partial (Homo sapiens) (accession number KAI2529781.1) with 11% query coverage, 32.08% identity and 47.8% similarity. Hyaluronidases are enzymes that break the glycosidic bond between glucuronic acid and N-acetylglucosamine to degrade hyaluronic acid, a significant component of the extracellular matrix. Despite playing a crucial role in the dissemination and toxicity of venom, snake venom hyaluronidases (svHA) have received less attention in comparison to exploring the human hyaluronidase enzyme.53 Human and snake venom hyaluronidases both have conserved amino acid residues in the catalytic domain. The catalytic domain of hyaluronidases is classified as a glycoside hydrolase family 56 (GH56) domain and has a characteristic (β/α) 8 barrel fold. A non-catalytic linker found in the link domain of human hyaluronidase (hHA) is thought to be involved in protein-protein interactions and the regulation of enzyme activity. However, snake venom hyaluronidases are simple molecules and do not possess a separate link domain, but the catalytic domain contains surface-exposed loops that may participate in substrate binding. Some svHA also contain a C-terminal disintegrin-like domain, such as the hyaluronidase from the venom of the snake Bothrops asper, which may be involved in binding to integrins and other extracellular matrix proteins. Despite the similarities between the catalytic domains of human and snake venom hyaluronidases, there are also differences in their biochemical properties and substrate specificities.54,55 For example, snake venom hyaluronidases often exhibit broader substrate specificity, degrading not only hyaluronic acid but also chondroitin sulfates, whereas human hyaluronidases are more selective. Additionally, venom hyaluronidases are generally more heat-stable and function effectively across a wider pH range compared to their human counterparts.
Figure 6. Sequence alignment for the snake venom Hyaluronidase and the corresponding human protein, which has the most similarity. In this figure, the protein with UniProt ID V8PG63 represents a snake venom hyaluronidase of Ophiophagus hannah species, while KAI2529754.1 corresponds to a human hyaluronidase protein
L-amino oxidase
Sequence analysis indicates that L-amino oxidase isoform 1 precursor and interleukin 4 induced 1 of Homo sapiens (accession number NP_690863.1) has more sequence similarity with snake venom L-amino oxidase of Ophiophagus hannah species (UniProt accession number P81383) with 96% query coverage, 37.17% identity and 56.5% similarity (Figure 7). Although the alignment with peroxisomal N(1)-acetyl-spermine/spermidine oxidase isoform 4 (Homo sapiens) (NP_997011.1) shows relatively high similarity (64.2%) within the aligned region, it spans only 56% of the query sequence. Snake venom LAAOs are typically monomeric proteins with molecular weights ranging from 50-140 kDa. They are known to contain two or more domains. N-terminal flavin adenine dinucleotide (FAD)-binding domain and a C-terminal catalytic domain are the two constitutive domains in these proteins. The catalytic domain is more variable and can vary in length and sequence composition, whereas the FAD-binding domain is highly conserved and crucial for enzyme function. Some snake venom LAAOs also contain additional domains, such as a C-type lectin-like domain or a disintegrin-like domain, which may contribute to their distinct biological activity and toxicity. LAAOs in humans are expressed in a variety of tissues, including the placenta, kidney, and liver. The molecular weight of human LAAOs is approximately 140 kDa and is a homo-dimeric protein. They contain three domains: N-terminal signal peptide, a middle domain, and a C-terminal FAD-binding domain. The middle domain is more variable and contributes to the substrate specificity and impacts the catalytic efficiency of the enzyme.15
Figure 7. Sequence alignment for the snake venom L-amino acid oxidase (LAAO) and the corresponding human protein, which has the most similarity. In this figure, the protein with UniProt ID P81383 represents a snake venom L-amino oxidase of Ophiophagus hannah species, while AAH64378.2 corresponds to a human L-amino acid oxidase protein
The structural and functional similarities between venom proteins and human counterparts underpin their dual applications in venomics. The resemblance of venom proteins to human enzymes and receptors enables their direct application in drug development. For instance, PLA2‚ interaction with lipid membranes inspires anti-inflammatory and anti-cancer strategies. Understanding these similarities helps predict venom protein interactions in the human body, enabling targeted inhibition of toxic effects while preserving therapeutic potential. By addressing these gaps, venomics can fully leverage the similarities between snake venom and human proteins to optimize drug discovery and improve antivenom therapies. While many venom proteins show promise as drug candidates, their inherent toxicity issues limit their clinical application. There is a lack of detailed structure-function studies and scalable production methods for venom-derived drugs. Traditional antivenoms are broad-spectrum but lack efficacy against specific venom components. There is limited integration of venomics data into antivenom production to achieve species-specific or toxin-specific efficacy.
Harnessing Snake venom proteins for biomedical breakthroughs
Interestingly, the same properties that make venom harmful in envenomation hold significant promise for developing innovative therapies. Snake venom proteins offer significant benefits due to their relatively low molecular mass. Their compact size makes them easier to be synthesized, both biologically and chemically, allowing for large-scale production. The simple structure of these proteins, in terms of their secondary and tertiary conformations, facilitates a clearer understanding of their roles and interactions, making them valuable templates for drug design. Furthermore, the flexible loops within snake venom proteins grant those unique biological activities and therapeutic potential. These loops can interact with a wide range of targets, adding versatility and efficacy, and they can be customized for specific binding with high precision, making them useful for molecular recognition and modulation. Since these proteins lack complex post-translational modifications, production, characterization, and application are simplified, reducing both complexity and costs. This simplicity also makes it easier to study their structure-function relationships and biological activities, offering insights into their mechanisms of action. There has been a mixed information in literature regarding the thermodynamic stability of the snake venom proteins. Such instability also has been pointed out to be a deterrent for the production of snake antivenom. Therefore, a clear investigation on the thermodynamic stability of different snake venom proteins should be taken up. This necessitates the production of mutant proteins with significant stability. Stable proteins are more likely to behave consistently and predictably, which is crucial for drug discovery and diagnostic applications.
Snake venom proteins for human therapeutics
Antibacterial activity
Toxins from various Viperid and Elapid species exhibit bactericidal effects. The basic PLA2‚ extracted from Bothrops marajeonsis showed no inhibitory effect against Pseudomonas aeruginosa or Staphylococcus aureus,56 while the acidic PLA2‚ isolated from Bothrops erythromela inhibits the growth of Gram-positive bacteria but does not affect Gram-negative bacteria.57 In contrast, basic PLA2‚ derived from Daboia russellii pulchella demonstrated enhanced bactericidal activity against Gram-positive bacteria, while showing less impact on Gram-negative bacteria.58 The bactericidal effect of PLA2, especially the basic form, has been linked to its ability to disrupt bacterial membrane integrity.59 Although catalytic activity is not the sole determinant of bactericidal effect, there appears to be a correlation between hemolytic and bactericidal activity of D. russellii pulchella PLA2.58 Additionally, p-bromophenacyl bromide (p-BPP) not only helps in reducing enzyme activity but also helps in destabilizing the cell wall of the bacteria.59
Crotoxin A and B (PLA2‚ -CA and PLA2‚ -CB) from Crotalus durissus terrificus exhibit antibacterial activity against Ralstonia solanacearum.60 Likewise, crotoxin B from C. durissus terrificus and daboiatoxin from Daboia russelli show inhibitory effect against two strains of Burkholderia pseudomallei (TES and KHW).61 While some PLA2‚ variants display bactericidal activity against various range of bacteria, including Gram-positive and Gram-negative.62,63 others specifically target Gram-positive bacteria.64 This suggests a complex relationship between the bactericidal effects of PLA2‚ and its sequence and structure.
L-amino acid oxidases (LAAOs) also exhibit antibacterial activity.56,65-68 It has been reported that Bothrops leucurus venom can cause a dose-dependent inhibition of S. aureus growth, with a MIC value of 25 µg/ml. SvLAAOs from Crotalus adamanteus and Bothrops asper show antibacterial activity against S. aureus and Proteus mirabilis, similar to SvLAAOs from Bothrops venoms.56,69,70 Additionally, Pseudomonas aeruginosa and Escherichia coli growth are inhibited by svLAAO from Bothrops pirajai venom.70 Peptides derived from svLAAO showed enhanced antibacterial effects compared to the whole protein, suggesting that smaller peptide fragments may be promising candidates for novel peptide-based antibiotics.71 The bactericidal mechanism of svLAAOs is generally attributed to the induction of oxidative stress in bacterial cells, leading to disruption and increased permeability of the plasma membrane, ultimately causing cell death. This process is associated with the presence of hydrogen peroxide in the reaction medium.72
The three-finger toxin (3FTx) family member, gamma toxin from the cobra Naja nigricollis venom, increases the membrane permeability of both Gram-positive and Gram-negative bacteria, causing bactericidal effects. Its physical interaction with primary membrane proteins demonstrates direct molecular activity on the components of Gram-negative and Gram-positive bacteria, namely lipopolysaccharide (LPS) and lipoteichoic acid (LTA), respectively. Disrupting the LPS layer and inhibiting LTA production leads to bactericidal effects.73 Cardiotoxin 3 from venom of Naja naja atra, another 3FTx family member, shows a similar mode of action but is more effective against S. aureus than E. coli.74
Lectins and their homologs, like BlL from Bothrops leucurus venom, also have antibacterial properties. BlL and a protein from Bothrops jararacussu venom inhibit S. aureus by interfering with biofilm formation.75 The Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) for S. aureus are 31.5 µg/ml and 500 µg/ml, respectively, while for Bacillus subtilis, the corresponding values are 125 µg/ml and 250 µg/ml.76
Antimicrobial peptides (AMPs) are a critical part of the innate immune defense on human skin. Two main categories of AMPs found in humans are defensins and cathelicidins, many of which are effective against Staphylococci.77 These peptides help to eliminate bacteria from neutrophil phagosomes through phagocytosis. Like humans, snakes also produce cathelicidins, which are smaller cationic AMPs (cAMPs). These peptides are highly effective against various bacteria, enveloped viruses, and fungi.78 Ophiophagus hannah cathelicidin (OH-CATH) and its analogues exhibit potent antibacterial and moderate hemolytic effects.79 They are more effective than commonly used natural antibiotics against Acinetobacter species, including multi-drug-resistant Acinetobacter baumannii (MRAB) and methicillin-resistant S. aureus (MRSA).80 AMPs rich in cysteine demonstrate broad antibacterial activity, with their positively charged surface and flexible shape facilitating interaction with the negatively charged components of bacterial membranes, thus enhancing their antibacterial effects.78 CoaTx-II, a small peptide derived from Crotalus oreganus abyssus based on the primary structure of Lys49 PLA2, shows antibacterial activity against drug-resistant clinical isolates. The peptide’s charged and aromatic amino acids are crucial for its interaction with bacterial cell membranes81 (Table 1).
Table (1):
Antibacterial activity of snake venom protein against different bacteria with their proposed active components
| Snake Venom protein | Species | Antibacterial Component | Effective against | Ref. |
|---|---|---|---|---|
| L-amino oxidase | B. leucurus | BleuLAAO | S. aureus | 128 |
| Agkistrodon halys Pallas | LAAO | E. coli K12D31 | 129 | |
| B. jararaca | LAAO | S. aureus | 130 | |
| B. marajoensis | BmarLAAO | S. aureus and P. aeruginosa | 128 | |
| T. jerdonii | TJ-LAO | E. coli, S. aureus, P. aeruginosa, and B. megaterium | 131 | |
| Trimeresurus Mucrosquamatus Agkistrodon | TM-LAAO | E. coli, S. aureus and B. dysenteriae | 132 | |
| blomhoffii ussurensis | Akbu-LAAO | S. aureus | 133 | |
| Bothrops Mattogrossensis | BmLAAO | Gram-positive and -negative bacteria | 134 | |
| Ophiophagus hannah | King cobra L-amino acid oxidase (Oh-LAAO) | Gram-positive and -negative bacteria | 135 | |
| B. alternatus | Balt-LAAO-I | E. coli and S. aureus | 135 | |
| Daboia russellii siamensis | DRS-LAAO | S. aureus (ATCC 25923), P. aeruginosa (ATCC 27853) and E. coli (ATCC 25922) | 66 | |
| King cobra venom | LAAO | S. aureus, S. epidermidis, P. aeruginosa, K. pneumoniae, and E. coli | 136 | |
| Naja naja oxiana C. durissus | LAAO | B. subtilis and E. coli | 65 | |
| Cumanensis | CdcLAAO | S. aureus and A. baumannii | 71 | |
| Porthidium nasutum | PnPLA2 | S. aureus | 137 | |
| PLA2 | Bothrops asper | PLA2 myotoxins | S. typhimurium and S. aureus | 138 |
| Vipera berus berus | VBBPLA2 | B. subtilis | 139 | |
| Echis carinatus | EcTx-I | E. aerogenes, E. coli, P. vulgaris, P. mirabilis, P. aeruginosa and S. aureus | 140 | |
| Agkistrodon spp. | AgkTx-II | S. aureus, P. vulgaris and B. pseudomallei | 141 | |
| Bungarus fasciatus | BFPA | E. coli and S. aureus | 67 |
Analgesic and antinociceptive activity
Controlling pain with the best and most appropriate drugs is still a challenge in healthcare systems.82 Researchers have discovered that PLA2‚ derived from snake venom exhibits efficacy in alleviating pain and inflammation in animal models experiencing arthritis and neuropathic pain.83 Distinct mechanism of action and potent analgesic effects exhibited by the isolated neurotoxin, PLA2‚ and myotoxin from snake venom make them potential therapeutic drugs for pain treatment.84 Several studies have highlighted the analgesic potential of various snake venom toxins. For example, κ-bungarotoxin has been shown to exhibit significant analgesic effects in animal models.85 Crotoxin has also demonstrated efficacy in treating neuropathic pain and inflammation.55 These peptides work by rapidly and irreversibly inhibiting specific subtypes of neuronal acid-sensing ion channels (ASICs) found in the central nervous system (CNS).86 ASICs are typically activated by protons, which induce pain sensations in a reversible manner. By blocking these channels, these toxins provide an analgesic effect in both acute and chronic inflammatory pain conditions.87
Anti-arthritic and anti-inflammatory activity
Various animal models are being utilized to examine the pathophysiological effects of snake venom toxins on arthritis. For instance, the venom of Naja kaouthia has been shown to induce significant changes in arthritis biomarkers in mouse models, including alterations in paw and ankle diameter, urinary markers such as hydroxyproline and glucosamine, biochemical markers like acid and alkaline phosphatase, the molecular marker IL-10, and liver antioxidant parameters such as catalase and glutathione.88 Additionally, the anti-arthritic and anti-inflammatory effects of the NN-32 peptide, derived from Indian cobra venom, were evaluated using mouse models of Freund’s complete adjuvant (FCA)-induced arthritis and carrageenan-induced inflammation. The results demonstrated that NN-32 peptide treatment significantly reduced physical and urinary parameters, serum enzyme levels, and cytokine levels compared to the arthritic control group. Furthermore, NN-32 peptide treatment effectively alleviated carrageenan-induced inflammation in rats, suggesting that this cytotoxic NN-32 protein possesses both anti-arthritic and anti-inflammatory properties.89
The prolonged anti-inflammatory effects observed with crude Crotalus durissus terrificus venom are attributed to crotoxin, an isoform of PLA2‚ by activating the formyl peptide receptors, which acts as a major factor in enhancing the anti-inflammatory effects.76 In a related study, a single dose of Crotalus durissus terrificus venom (CdtV) results in prolonged effect of altering the inflammatory response with changes in the major symptoms such as paw edema and migration of the cells observed in the mice, where inflammation is induced by carrageenan compound similar to the effects observed with the crude venom.90
Anti-cancer activity
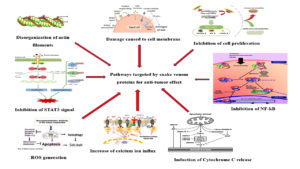
Multiple research has stated that snake venom protein PLA2‚ and LAAO possess anti-cancer property by inducing apoptosis mechanism leading to cell death, altering the cell cycle, reducing the cell growth to G0 and G1 phase, and further stops the proliferation of the cells, thereby helping in controlling the cancer metastasis in the body. Both apoptosis and necrosis are observed in cells treated with these enzymes. The cytotoxicity of PLA2 is mainly attributed to its C-terminal region, which interacts with the cell membrane. In contrast, LAAO produces significant amounts of H2O2 through its enzymatic activity, leading to cell death by accumulating the reactive oxygen species (ROS) in the cells. Along with LAAO, some disintegrins are also known to induce apoptosis. The disintegrin derived from Naja naja venom has been reported to suppress the proliferation of MCF-7 (IC50 = 2.5 ± 0.5 µg/mL), A549 (3.5 ± 0.5 µg/mL), and HepG2 (3 ± 0.5 µg/mL) cell lines.91 Additionally, some compounds extracted from the snake venom toxins are reported to possess anti-melanoma activity. The possible mechanism of action for anti-melanoma activity might be because of the highly specificity of the disintegrins for disrupting the integrity integrins.92
PLA2‚ and LAAO also demonstrate anticoagulant activity, presenting potential applications in cancer treatment, particularly in addressing venous thromboembolism frequently seen in cancer patients. Nevertheless, the precise mechanism by which these enzymes induce cell death remains unclear, particularly their interactions with death receptors, is not well understood and requires further investigation. Ensuring the selectivity of enzymes towards cancer cells is essential, as non-cancer cells are less sensitive to their cytotoxic effects. These enzymes exhibit stronger cytotoxic activity in cancer cells than in normal cells, making them promising candidates for potential use as chemotherapeutic agents. It is believed that these enzymes promote oxidative stress. PLA2‚ during lipolysis, can generate ROS, while LAAO produces H2O2 both of which contribute to cell death13 (Table 2, Figure 8).
Table (2):
Anticancer agents from snake venom proteins
No. |
Snake venom component – (UniProt ID) |
Snake species |
Mechanism of Action |
Ref. |
|---|---|---|---|---|
1. |
Phospholipases A2 (P0CAS0) |
Cerastes cerastes |
It acts specifically on integrins aβ and a5β1 and has antiangiogenic and anticancer activities |
142 |
2. |
Hyaluronidase (P86100) |
Mesobuthus martensi |
Degrades hyaluronan and alters the expression of a CD44 isoform in breast cancer cells |
143 |
3. |
L-amino acid oxidase (P81383) |
Ophiophagus hannah |
Reduces cell proliferation and declines thymidine therefor decreases the uptake of thymidine in murine fibrosarcoma, melanoma, and colorectal cancer |
13 |
4. |
Cathelicidin-BF (B6D434) |
Bungarus fasciatus |
Cell Proliferation of B16F10 cell line and B16 cell line is inhibited |
144 |
5. |
Disintegrin (P0C6S4 – Predicted) |
Vipera xanthine |
Prevents tumor cell invasion and inhibits OVCAR-5 cell attachment to extracellular matrix proteins |
145 |
6. |
Viperistatin (P0C6E2- Predicted) |
Vipera palaestinae |
The engagement of integrins with the extracellular matrix is crucial for the migration and invasion of cancer cells; disrupting this interaction hinders both their adhesion and movement. |
146 |
7. |
Colombistatin (P18618- Predicted) |
Bothrops colombiensis |
Inhibition of ADP-induced platelet aggregation |
65 |
8. |
BJcuL (lectin) (P83519- Predicted) |
Bothrops jararacussu |
Inhibits tumor progression and endothelial cell proliferation, while also promoting erythrocyte agglutination |
147 |
9. |
Obtustatin (P83469- Predicted) |
Vipera lebetina obtuse |
It possesses antiangiogenic activity |
148 |
10. |
Lebestatin (Q3BK14- Predicted) |
Macrovipera lebetina |
Specifically binds to the α1β1 integrin found on laminin-1 and collagen. The interaction between integrins and the extracellular matrix is vital for cancer cell adhesion and migration |
149 |
11. |
Rhodostomin (P30403- Predicted) |
Agkistrodon rhodostoma |
Disrupts the binding between integrins and extracellular matrix proteins, an interaction essential for cancer cell proliferation |
150 |
12. |
Batroxobin (P04971) |
Bothrops atrox |
Inhibits the differentiation and metastatic progression of cancer cells |
151 |
13. |
Snake venom toxin (Q9PT41) |
Vipera lebtina turanica |
Restricts the proliferation of cancer cells by inducing cell cycle arrest at the G2-M phase. It also suppresses NF-κB, a key anti-apoptotic transcription factor, and significantly inhibits the nuclear translocation of the p50 subunit |
152 |
14. |
Saxatilin (Q9DGH6) |
Gloydius saxatilis |
Decreased cell invasion through MMP-9 regulation function in MDAH 2774 |
153 |
15. |
Lebectin (W5XDMO) |
Macrovipera lebetina |
Impedes tumor cell adhesion, migration, and invasion, while also inhibiting angiogenesis |
154 |
16. |
Eristostatin (P0C6S4) |
Eristicophis macmahoni |
Suppresses the establishment of melanoma cells in lung and liver tissues |
155 |
17. |
Ammodytoxin-C (P11407) |
Vipera ammodytes |
Induces autophagy by enhancing autophagosome formation and simultaneously promotes apoptosis |
156 |
18. |
Crotatroxin 2 (P68520) |
Crotalus atrox |
Prevents fibrinogen from binding to GP IIb/IIIa receptors, while also inhibiting cell migration and tumor colonization |
157 |
19. |
Α-Bungarotoxin (P60615) |
Bungarus multicinctus |
Exerts antiproliferative effects on cancer cells by inducing G1 phase arrest and counteracting nicotine-induced NK cell-mediated proliferation |
158 |
20. |
MVL-PLA2 (B5U6Z2) |
Macrovipera lebetina |
Inhibits angiogenesis and triggers alterations in the actin cytoskeleton |
159 |
21. |
Dendroptoxin-ĸ |
Dendroaspis polylepis |
Demonstrates antitumor activity by disrupting the G1 to S phase transition, increasing the expression of p27Kip1, p21Waf1/Cip1, and p15INK4B, and inhibiting cyclin-dependent kinases (CDKs) involved in cell cycle regulation |
160 |
22. |
Contortrostatin (Q9IAB0) |
Agkistrodon controtrix |
Blocks platelet aggregation and inhibits cancer cell growth, adhesion, migration, and the formation of new blood vessels |
161 |
23. |
Mojastin1 (P0C7X7) |
Crotalus scutulatus |
Interferes with ADP-induced platelet aggregation and suppresses cell migration, invasion, and tumor establishment |
161 |
24. |
Leucurogin (P0DJ87) |
Bothrops leucurus |
It possesses antiangiogenic activity |
162 |
25. |
Leucurolysin-B (P86092) |
Bothrops leucurus |
Demonstrates cytotoxic effects against a range of cancer cell lines, including U87, T98, MCF7, RT2, EAC, and UACC. |
163 |
26. |
Vicrostatin |
Echis carinatus |
Limits the migration potential of human umbilical vein endothelial cells (HUVECs) |
164 |
27. |
Pooled venom (A0A7R7T1Q6) |
Montivipera Xanthine |
Displays dose-dependent cytotoxicity and inhibits cell proliferation |
165 |
28. |
Viridistatin 2 (A2CJE6) |
Crotalus viridis |
Suppresses proliferation, migration, adhesion, and survival of human pancreatic carcinoma (BXPC-3) cells by inducing apoptosis |
166 |
29. |
Tzabcanin (C0HK50) |
Crotalus tzabcan |
Prevents the adhesion of melanoma (A-375) cells and lung carcinoma (A-549) cells to vitronectin |
167 |
30. |
Ophiophagus hannah venom (12C090) |
Ophiophagus hannah |
Showed inhibitory properties on tumour cell induced angiogenesis |
168 |
Anti-viral activity
Recent studies have highlighted the antiviral effects of various snake venom components.93,94 Non-cytotoxic fractions of Cdt venom have shown anti-viral activity against the measles virus, inhibiting its replication in Vero cells at concentrations of 0.1 µg/mL and 100 µg/mL.95 Naja nigricollis venom has demonstrated in controlling the viral load in the human erythrocytes which are infected with the Sendai virus. Virus-infected cells exhibited tenfold higher susceptibility, by lysing the two among five venom toxins selected for the study, with 4 identified cytotoxins from Naja nigricollis venom showing that virus-infected cells were ten times more vulnerable to cytotoxic effects than healthy cells.93 Additionally, LAAO from Bothrops jararaca has been shown to inhibit the viral growth of dengue virus, therefore possessing antiviral properties. Another study states that the cells infected with dengue type 3 virus (DENV-3) had reduced viral load after treatment with LAAO compared to untreated cells.70
The venom of Naja siamensis contains an oxidized derivative of alpha-toxins and immunokines, which suppresses the lymphocytes infection caused by HIV and Feline Immunodeficiency Virus (FIV). There is a significant resemblance between the amino acid sequences of long-chain neurotoxins protein present in Naja siamensis and Bungarus multicinctus venoms and a short sequence in the HIV-1 gp120, suggesting that these molecules may compete for the same binding sites.96,97 Additionally, a metalloprotease inhibitor isolated from Trimeresurus stejnegeri venom has the potential to block protease enzymes and prevent the formation of new virus particles.
PLA2‚ from snake venom prevents HIV- 1 replication in primary human leukocytes by blocking viral entry before the virus uncoats and releases proteins from the viral capsid.98,99
LAAO from venoms of Crotalus atrox, Pseudechis australis, and Trimeresurus stejnegeri showed dose-dependent inhibition of HIV-1 infection and replication by blocking the p24 antigen.44,100,101 Free radicals like H2O2, produced during LAAO activity, contribute to the antiviral effects by preventing HIV infection and replication, though H2O2 interaction with catalases may reduce its antiviral activity.101 Snake venoms from Crotalus adamanteus, Oxyuranus microlepidotus, Bungarus candidus, Hydrophis cyanocinctus, Naja naja, Notechis ater, Naja sumatrana, and Naja kaouthia have also been reported to exhibit anti-HIV activity.61,102,103
HIV entry requires CD4 and a coreceptor, primarily CCR5 or CXCR4, which determine viral tropism and disease progression. Discoveries of coreceptor roles and CCR5 mutations have provided key insights into HIV biology and therapeutic targets.104
PLA2‚ from snake venom prevents HIV-1 replication in primary human leukocytes by blocking viral entry before the virus uncoats and releases proteins from the viral capsid.98,99,104 Anti-HIV activity has been demonstrated by synthetic peptides derived from PLA2‚ including p3bv peptide, which blocks HIV-1 attachment to T cells by binding to the CXCR4 receptor, thereby preventing virus entry.105 Crotoxin, a phospholipase isolated from Crotalus durissus terrificus venom, has been shown to inhibit HIV in vitro.106 Chemokines and their derivatives can compete with the HIV-1 gp120 for binding to receptors like CXCR4, effectively suppressing HIV replication.104,107
PLA2 from snake venom has antiviral potency against SARS-CoV-2, particularly the dimeric form. On the other hand, dimeric PLA2 catalytic activity determines its virucidal and antiviral properties. The unique ability of PLA2 is to inactivate the virus from spreading infection.108 Batroxobin is a safe drug that is used extensively for perioperative bleeding and is effective for a variety of illnesses, such as deep vein thrombosis and pulmonary embolism.109 This provides the opportunity to introduce Batroxobin, which is isolated from Bothrops atrox venom and catalyzes the conversion of fibrinogen to fibrin (Pefakit Reptilase Time; Pentapharm), as part of an anti-COVID-19 therapeutic strategy aimed at preventing the deadly pulmonary embolism that can lead to severe SARS-CoV-2 infections.110
Obstacles encountered in the drug development journey
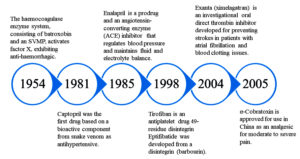
The most common application of animal toxins is as pharmacological tools for target validation. Although there have been many instances of success, the availability of authorized compounds possessing significant pharmacological properties derived from animal venoms remains limited. Challenges in developing animal toxin-based drugs are commonly linked to gaps in fundamental research, preclinical assessment, and clinical trials (Figure 9). Additionally, a lot of the difficulties encountered during the different phases of drug development are not been sufficiently documented in the scientific literature because the information about compounds that are in the development stage and for which the development is halted due to many internal problems, as they are protected by intellectual property rights.111
Figure 9. Chronological overview of Food and Drug Administration (FDA) approved drugs derived from snake venom toxins
During the initial phase of research
Studies utilizing animal toxins are not simple tasks because several concerns need to be overcome. Genomic analysis and cDNA library are the choices which can be explored to strengthen the research.112 Recent studies have shown that these toxic molecules can be obtained by utilising organoid technology. However, this is still a challenging and daunting task.113
Second phase of research (preclinical phase)
The pharmaceutical applications of drugs formulated from snake venom toxins have been extensively investigated due to their diverse range of biological activities. These toxins showed shown significant activity in areas such as anticoagulation, anti-thrombosis, antimicrobial activity, analgesic properties, and potential for anti-tumour treatments. However, several limitations and challenges need to be resolved before using snake venom toxins as drugs. Selectivity, mode of action, formulation, stability, and cost of production are some constraints with toxin-based drugs.114 The selectivity of snake venom toxins may have off-target effects due to their non-specific binding to other proteins and enzymes, which can result in unwanted side effects. Therefore, the development of toxin-based drugs is a major challenge for achieving high selectivity for the target protein or enzyme. Understanding the mechanism of action of toxin-based drugs is vital in developing pharmaceuticals reliant on their biological activity. Nevertheless, it is challenging to determine the exact mechanism of action and the complexity of the interactions between toxins and their target molecules, which often requires extensive research and experimentation. Another challenge in the development of drugs based on snake venom toxins is that the size of the molecules is often large, complex, and unstable. However, compared to humans, these have better physicochemical properties. The purification, storage, and delivery of these drugs entail a multifaceted process that demands meticulous attention to factors like pH, temperature, and stability. The stability of snake venom toxins is a crucial factor in the development of drugs based on toxins. These molecules limit their therapeutic efficacy unless targeted delivery systems are chosen, are prone to degradation and can be rapidly eliminated from the body. Therefore, appropriate drug delivery systems should be developed. The cost of production of snake venom toxin-based drugs is also a major challenge. The extraction and purification process of toxins is both time-consuming and expensive, making it challenging to produce these molecules on a large scale, often rendering it infeasible. Purification also introduces complexity, as many of the proteins are bound to each other, especially when they are being purified under native conditions. Cloning and expression of certain snake venom proteins can help to overcome the problems associated with purification.115,116 Despite the obstacles faced, there have been notable advancements in drug development through the utilization of snake venom toxins. For example, the snake venom derived Echistatin,117 has been used as an antiplatelet agent, and extensive research has been conducted on crotoxin, a constituent extracted from the venom toxins of the South American rattlesnake Crotalus durissus terrificus, because of its potential antitumor properties. To advance the formulation of snake venom toxin-oriented drugs and enhance their effectiveness and safety, additional research is required to tackle the associated challenges.21
Final phase of research (clinical trial)
The standard method for assessing certain drug-related issues involves assessing the efficacy, safety and the long-term effects. However, at times the studies lack access to specific population, especially children, pregnant women and elderly people.118 Therefore, the developmental process needs to address these constraints and identify which drugs need more epidemiological research. Electronic health care records for post-marketing and comparing the safety of the medication can be utilized to overcome these limitations.119 In addition, snake venom toxin-based drugs face regulatory challenges, particularly in countries where there is limited experience with these types of drugs. Moreover, the regulatory process for approving drugs is lengthy and not economical. There are also ethical considerations related to the applications of snake venom toxins in clinical trials, and obtaining venom, even for research purposes, is becoming a cumbersome process. The challenge also includes concerns about exposing patients to unnecessary risks or about the use of animal testing in the formulation of new and suitable drugs.120 Due to these constraints, despite the high therapeutic potential of snake venom proteins, the transition from the laboratory to bedside is very slow and challenging. Drugs, which are in the phase of trials and being approved by the Food and Drug Administration (FDA) are listed in Table 3.
Table (3):
Food and Drug Administration (FDA) approved snake venom-based drugs
| Approved drugs | Drugs | Venomous Snake | Mode of action | Medical Treatment | Production | Ref. |
|---|---|---|---|---|---|---|
| FDA approved | Captopril (Capoten®) Hemocoagulase | Bothrops jararaca Bothrops atrox | Inhibiting ACE Coagulation of the blood is catalyzed | Hypertension, cardiac failure Abdominal surgery, human vitrectomy, and plastic surgery | Synthetic Purified from venom | 169, 170, 111, 171 |
| Eptifibatide (Integrilin®) | Sistrurus miliarus barbouri | Inhibits the platelet aggregation by preventing ligand binding to Glycoprotein (GP) IIb/IIIa platelet receptor | Coronary syndrome, acute | Synthetic | 172, 173 | |
| Tirofiban (Aggrastat®) | Echis carinatus | Acts as an antagonist by preventing fibrinogen from binding to the GPIIb/IIIa receptor on platelets | Heart attack | Synthetic | 174-176 | |
| Batroxobin (Defibrase®, Plateltex-Act®, Vivostat®) | Bothrops atrox & B. moojeni | Cleaves alpha-chain of fibrinogen | Associated with conditions such as deep vein thrombosis, stroke, pulmonary embolism, and myocardial infarction | Purified from venom | 171, 177 | |
| Exanta (Ximelagatran) | Cobra venom | Inhibiting direct thrombin | Thromboembolic side effects associated with atrial fibrillation | Synthetic | 178 | |
| Cobratide (Ketonging, cobrotoxin) | Naja naja atra | Inhibits the activity of nicotinic acetylcholine receptors | Chronic arthralgia, sciatica, neuropathic headache | Purified from venom | 84 |
Snake venom proteins represent a promising candidate for human therapeutics, with active compounds showing significant effects in treating conditions such as cardiovascular diseases, cancer, and pain management. Notable compounds include Bradykinin Potentiating Peptides and snake venom metalloproteinases, which are being explored for their medicinal properties.
Snake venom proteins offer significant benefits due to their relatively low molecular mass. Regulatory challenges and scalability issues further hinder progress. Integrating nanotechnology into this process offers promising solutions, enabling improved drug delivery, enhanced stability, and targeted action while minimizing side effects. Nanocarriers such as liposomes, nanoparticles, and dendrimers can optimize venom-derived therapeutics, overcoming conventional barriers. This innovative approach not only enhances efficacy but also reduces the risks associated with venom-based drugs, marking a significant advancement in addressing critical challenges in modern drug development.
Integrating nanotechnology in drug development
Nanotechnology has become a promising approach in the development and formulation of drug, offering improved effectiveness and reduced adverse effects when compared to conventional therapies. Through use of nanoscale drug delivery systems, nanoparticle technology alters the kinetics, distribution, and release of drugs, providing several benefits, such as increased patient compliance, lower healthcare costs, and enhanced drug efficacy while reducing toxicity.121 Various nanoparticle formulation approaches have been investigated to improve the targeted delivery of venom peptides. This approach is gaining momentum due to its ability to reduce systemic toxicity and improve therapeutic efficacy. For instance, the use of nanoparticle systems that efficiently destroy tumor cells directed by immune cells to play an important role in preventing the tumors development. Recent studies have focused on the development and exploration of nanoparticle-conjugated venom peptides to enhance therapeutic efficacy and reduce toxicity.122
Establishing the efficacy, potency, and safety of enzymatic toxins is crucial before they can be developed into chemotherapeutic agents. Targeted delivery approaches, such as nanoparticles formulation or conjugating with the ligands or production of monoclonal/polyclonal antibodies, need to be explored to recognize cancer cells. NN-32 purified toxin and toxin conjugating with nanogold GNP-NN-32 demonstrates remarkable cytotoxic potential against MCF-7 and MDA-MB-231 cell lines, effective interaction with dose and duration exposure.123
Formulation of gold nanoparticles with Naja kaouthia cytotoxin 1 (NKCT1) have a synergistic effect that reduces the required dosage and course of action of the NKCT1. This controlled release of NKCT1 to target cells via GNP also increases the cytotoxic effect by two to threefold and minimizes the toxic effects of NKCT1. Due to its ability to induce cell cycle arrest, promote apoptosis, and regulate nuclear fragmentation, conjugation has shown high antileukemic action. GNP-NKCT1 treatment supports the emerging concept that conjugating nanoparticles may prove advantageous for leukemia, as GNP-NKCT1 regulates various biochemical pathways by inhibiting transcriptional and translational levels.124 Chitosan nanoparticles encapsulated with Echis carinatus snake venom result in increased efficacy compared to traditional adjuvant systems and also enhance the stability of the snake venom proteins in physiological systems.125 In another study, the apoptotic action of venom from Walterinnesia aegyptia (WEV) on breast cancer cells using free WEV and WEV formulated with silver nanoparticles has been able to inhibit the cell proliferation in a dose-dependent manner, with Walterinnesia aegyptia venom silver nanoparticle conjugate (WEV-NP) exhibiting greater efficacy and substantially enhancing the anticancer properties of WEV, according to WEV conjugated with silica nanoparticles (WEV-NP).126 In a recent study, CdtV (rattlesnake) was used to develop a fibrin sealant. This enzyme, which resembles thrombin, can convert fibrinogen into fibrin, which can naturally gel and is non-toxic, non-immunogenic, and biodegradable. For the formulation of fibrin sealant, multi-layered carbon nanotubes were utilized and nanohydroxyapatites for potential application in bone regeneration.127 The utilization of targeted delivery systems in employing snake venom toxins as therapeutic agents is crucial due to their ability to enhance specificity and minimize side effects on normal cells.
Snake venom research is advancing rapidly due to breakthroughs in genomics, proteomics, nanotechnology, and bioactivity assays, offering significant potential for therapeutic development. The structural and functional similarities between snake venom proteins and human proteins make them valuable models for designing enzyme inhibitors and drug candidates. These proteins exhibit diverse biological activities, supporting their use in treating various diseases. However, challenges remain in terms of ensuring target specificity, reducing toxicity without compromising function, delivering precise dosages, and scaling up production efficiently. Existing venom-derived drugs demonstrate that toxic components can be repurposed into safe and effective therapeutics. Moving forward, integrating bioinformatics for predictive modeling, enhancing delivery systems through nanotechnology, and deepening our molecular understanding of venom actions will be crucial to overcoming current limitations and translating venom-based research into innovative, clinically viable treatments.
ACKNOWLEDGMENTS
The authors would like to thank Vellore Institute of Technology, Vellore, Tamil Nadu, India, for the infrastructure support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PS conceptualized the study and wrote the original draft. PS and GJ reviewed the manuscript. GJ supervised the study. Both authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Chippaux JP. Snakebite envenomation turns again into a neglected tropical disease! J Venom Anim Toxins Incl Trop Dis. 2017;23:1-2.
Crossref - Gutierrez JM, Calvete JJ, Habib AG, Harrison RA, Williams DJ, Warrell DA. Snakebite envenoming. Nat Rev Dis Prim. 2017;3:17063.
Crossref - Togridou A, Graham SA, Owens JB, Santra V, Bharti O, Malhotra A. Prevention is better than cure: snakebite education in India. Education Sciences. 2019(4):75-96.
Crossref - Slagboom J, Kool J, Harrison RA, Casewell NR. Haemotoxic snake venoms: their functional activity, impact on snakebite victims and pharmaceutical promise. Br J Haematol. 2017;177(6):947-959.
Crossref - Tasoulis T, Isbister GK. A review and database of snake venom proteomes. Toxins (Basel). 2017;9(9):290.
Crossref - Munawar A, Ali SA, Akrem A, Betzel C. Snake venom peptides: Tools of biodiscovery. Toxins. 2018;10(110):474.
Crossref - Morsy MA, Gupta S, Dora CP, et al. Venoms classification and therapeutic uses: a narrative review. Eur Rev Med Pharmacol Sci. 2023;27(4):1633-1653.
- Oliveira AL, Viegas MF, da Silva SL, Soares AM, Ramos MJ, Fernandes PA. The chemistry of snake venom and its medicinal potential. Nat Rev Chem. 2022;6(7):451-469.
Crossref - Olaoba OT, Dos Santos PK, Selistre-de-Araujo HS, de Souza DHF. Snake venom metalloproteinases (SVMPs): a structure-function update. Toxicon: X, 7, 100052.
Crossref - Gutierrez JM, Escalante T, Rucavado A, Herrera C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins. 2016;8(4):93.
Crossref - Ferraz CR, Arrahman A, Xie C, et al. Multifunctional toxins in snake venoms and therapeutic implications: From pain to hemorrhage and necrosis. Front Ecol Evol. 2049;7:1-19.
Crossref - Ullah A, Masood R, Ali I, Ullah K, Ali H, Akbar H, Betzel C. Thrombin-like enzymes from snake venom: Structural characterization and mechanism of action. International journal of biological macromolecules. 2018;114:788-811.
Crossref - Hiu JJ, Yap MKK. Cytotoxicity of snake venom enzymatic toxins: Phospholipase A2 and L-amino acid oxidase. Biochem Soc Trans. 2020;48(2):719-731.
Crossref - Tan KK, Bay BH, Gopalakrishnakone P. L-amino acid oxidase from snake venom and its anticancer potential. Toxicon. 2018;144:7-13.
Crossref - Ullah A. Structure-Function Studies and Mechanism of Action of Snake Venom L-Amino Acid Oxidases. Front Pharmacol. 2020;11:1-17.
Crossref - Valentino PM, Boeno CN, Lopes JA, et al. Role of L -amino acid oxidase isolated from Calloselasma rhodostoma venom on neutrophil NADPH oxidase complex activation. Toxicon. 2018;145:48-55.
Crossref - Sridharan S, Kini RM, Richards AM. Venom natriuretic peptides guide the design of heart failure therapeutics. Pharmacol Res. 2020;155:104687.
Crossref - Kang TS, Georgieva D, Genov N, et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. FEBS J. 2011;278(23):4544-4576.
Crossref - Takeda S. ADAM and ADAMTS family proteins and snake venom metalloproteinases: A structural overview. Toxins. (Basel). 2016;8(5):8-11.
Crossref - Calvete JJ. Venomics: Integrative venom proteomics and beyond. Biochem J. 2017;474(6):611-634.
Crossref - Gutierrez JM, Lomonte B. Phospholipases A2: Unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon. 2013;62:27-39.
Crossref - Fox JW, Serrano SMT. Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8(4):909-920.
Crossref - Mackessy SP. Thrombin-like enzymes in snake venoms. In Toxins and hemostasis: from bench to bedside . Dordrecht: Springer Netherlands. 519-557:2010:
Crossref - Arlinghaus FT, Eble JA. C-type lectin-like proteins from snake venoms. Toxicon. 2012;60(4):512-519.
Crossref - Simoes-Silva R, Alfonso J, Gomez A, et al. Snake Venom, A Natural Library of New Potential Therapeutic Molecules: Challenges and Current Perspectives. Curr Pharm Biotechnol. 2018;19(4):308-335.
Crossref - Kini RM. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42(8):827-840.
Crossref - Montecucco C, Gutierrez JM, Lomonte B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell Mol Life Sci. 2008;65(18):2897-2912.
Crossref - Lomonte B, Rangel J. Snake venom Lys49 myotoxins: From phospholipases A2 to non-enzymatic membrane disruptors. Toxicon. 2012;60(4):520-530.
Crossref - Lomonte B, Gutierrez JM. Phospholipases A2 from viperidae snake venoms: How do they induce skeletal muscle damage? Acta Chim Slov. 2011;58(4):647-659.
- Lomonte B. Lys49 myotoxins, secreted phospholipase A2-like proteins of viperid venoms: A comprehensive review. Toxicon. 2023;224:107024.
Crossref - Lomonte B, Moreno E, Tarkowski A, Hanson LA, Maccarana M. Neutralizing interaction between heparins and myotoxin II, a lysine 49 phospholipase A2 from Bothrops asper snake venom. Identification of a heparin-binding and cytolytic toxin region by the use of synthetic peptides and molecular modeling. J Biol Chem. 1994;269(47):29867-29873.
Crossref - Nunez CE, Angulo Y, Lomonte B. Identification of the myotoxic site of the Lys49 phospholipase A2 from Agkistrodon piscivorus piscivorus snake venom: Synthetic C-terminal peptides from Lys49, but not from Asp49 myotoxins, exert membrane-damaging activities. Toxicon. 2001;39(10):1587-1594.
Crossref - Chioato L, Aragao EA, Ferreira TL, de Medeiros AI, Faccioli LH, Ward RJ. Mapping of the structural determinants of artificial and biological membrane damaging activities of a Lys49 phospholipase A2 by scanning alanine mutagenesis. Biochim Biophys Acta Biomembr. 2007;1768(5):1247-1257.
Crossref - Ward RJ, Chioato L, de Oliveira AHC, Ruller R, SA Juliana. M. Active-site mutagenesis of a Lys49-phospholipase A2: Biological and membrane-disrupting activities in the absence of catalysis. Biochem J. 2002;362(1):89-96.
Crossref - Conlon JM, Attoub S, Arafat H, et al. Cytotoxic activities of [Ser49]phospholipase A2 from the venom of the saw-scaled vipers Echis ocellatus, Echis pyramidum leakeyi, Echis carinatus sochureki, and Echis coloratus. Toxicon. 2013;71:96-104.
Crossref - Fox JW, Serrano SMT. Timeline of key events in snake venom metalloproteinase research. J Proteomics. 2009;72(2):200-209.
Crossref - Stocker W, Bode W. Structural features of a superfamily of zinc-endopeptidases: the metzincins. Curr Opin Struct Biol. 1995;5(3):383-390.
Crossref - Swenson S, Markland FS. Snake venom fibrin(ogen)olytic enzymes. Toxicon. 2005;45(8):1021-1039.
Crossref - Teixeira CFP, Landucci ECT, Antunes E, Chacur M, Cury Y. Inflammatory effects of snake venom myotoxic phospholipases A2. Toxicon. 2003;42(8):947-962.
Crossref - D’Amelio F, Vigerelli H, Prieto-da-Silva ARdB, et al. Bothrops moojeni venom and its components – an overview Fernanda. Toxins. 2021;13(7):26-33.
Crossref - Frost GI, Csoka T, Stern R, Yamagata S. The Hyaluronidases: A Chemical, Biological and Clinical Overview. Trends Glycosci Glycotechnol. 1996;8(44):419-434 (1996).
Crossref - Fox JW. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon. 2013;62:75-82.
Crossref - Wohlrab J, Finke R, Franke WG, Wohlrab A. Clinical trial for safety evaluation of hyaluronidase as diffusion enhancing adjuvant for infiltration analgesia of skin with lidocaine. Dermatologic Surg. 2012;38(1):91-96.
Crossref - Du XY, Clemetson KJ. Snake venom L-amino acid oxidases. Toxicon. 2002;40(6):659-665.
Crossref - Ali SA, Stoeva S, Abbasi A, et al. Isolation, structural, and functional characterization of an apoptosis-inducing L-amino acid oxidase from leaf-nosed viper (Eristocophis macmahoni) snake venom. Arch Biochem Biophys. 2000;384(2):216-226.
Crossref - Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19(16):4204-4215.
Crossref - Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: Classification and characterization. Biochim Biophys Acta. 2000;1488(1-2):1-19.
Crossref - Arni RK, Ward RJ. Phospholipase A2 – A structural review. Toxicon. 1996;34(8):827-841.
Crossref - Mounier CM, Ghomashchi F, Lindsay MR, et al. Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A2 occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A2-a. J Biol Chem. 2004;279(24):25024-25038.
Crossref - Castro-Amorim J, de Oliveira N, Da Silva SL, et al. Catalytically Active Snake Venom PLA2 Enzymes: An Overview of Its Elusive Mechanisms of Reaction. J Med Chem. 2023;66(1):5364-5376.
Crossref - Calvete JJ. Snake venomics: From the inventory of toxins to biology. Toxicon. 2013;75:44-62.
Crossref - Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4(8):617-629.
Crossref - Silva de Franca F, Tambourgi DV. Hyaluronan breakdown by snake venom hyaluronidases: From toxins delivery to immunopathology. Front Immunol. 2023;14:1-10.
Crossref - Girish KS, Kemparaju K. The magic glue hyaluronan and its eraser hyaluronidase: A biological overview. Life Sci. 2007;80(21):1921-1943.
Crossref - Boldrini-Franca J, Cologna CT, Pucca MB, et al. Minor snake venom proteins: Structure, function and potential applications. Biochim Biophys Acta Gen Subj. 2017;1861(4):824-838.
Crossref - Torres AFC, Dantas RT, Toyama MH, et al. Antibacterial and antiparasitic effects of Bothrops marajoensis venom and its fractions: Phospholipase A2 and L-amino acid oxidase. Toxicon. 2010;55(4):795-804.
Crossref - Bocian A, Slawek S, Jaromin M, et al. Comparison of methods for measuring protein concentration in venom samples. Animals. 2020;10(3):1-9.
Crossref - Sudarshan S, Dhananjaya, B. L. Antibacterial activity of an acidic phospholipase A2 (NN-XIb-PLA2) from the venom of Naja naja (Indian cobra). Springerplus. 2016;5:112.
Crossref - Correa EA, Kayano AM, Diniz-Sousa R, et al. Isolation, structural and functional characterization of a new Lys49 phospholipase A2 homologue from Bothrops neuwiedi urutu with bactericidal potential. Toxicon. 2016;115:13-21.
Crossref - de Cassia Alves R, Vieira Jr JR, Freire TC, et al. Snake venoms and purified toxins as biotechnological tools to control Ralstonia solanacearum. Pesqui Agropecu Bras. 2020;55:01756.
Crossref - Samy RP, Pachiappan A, Gopalakrishnakone P, et al. In vitro antimicrobial activity of natural toxins and animal venoms tested against Burkholderia pseudomallei. BMC Infect Dis. 2006;6:1-16.
Crossref - Samy RP, Kandasamy M, Gopalakrishnakone P, et al. Wound healing activity and mechanisms of action of an antibacterial protein from the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). PLoS One. 2014;9(2):e80199.
Crossref - Santamaria C, Larios S, Quiros S, et al. Bactericidal and antiendotoxic properties of short cationic peptides derived from a snake venom Lys49 phospholipase A2. Antimicrob Agents Chemother. 2005;49(4):1340-1345.
Crossref - Sudharshan S, Dhananjaya BL. Antibacterial potential of a basic phospholipase A2 (VRV-PL-VIIIa) from Daboia russelii pulchella (Russell’s viper) venom. Journal of Venomous Animals and Toxins including Tropical Diseases. 2015;21(1):17.
Crossref - Ciscotto P, de Avila RAM, Coelho EAF, et al. Antigenic, microbicidal and antiparasitic properties of an L-amino acid oxidase isolated from Bothrops jararaca snake venom. Toxicon. 2009;53(3):330-341.
Crossref - Vargas LJ, Quintana JC, Pereanez JA, Nunez V, Sanz L, Calvete J. Cloning and characterization of an antibacterial L-amino acid oxidase from Crotalus durissus cumanensis venom. Toxicon. 2013;64:1-11.
Crossref - Samel M, Tonismagi K, Ronnholm G, et al. L-Amino acid oxidase from Naja naja oxiana venom. Comp Biochem Physiol B Biochem Mol Biol. 2008;149(4):572-580.
Crossref - Phua CS, Vejayan J, Ambu S, Ponnudurai G, Gorajana A. Purification and antibacterial activities of an L-amino acid oxidase from king cobra (Ophiophagus hannah) venom. Journal of Venomous Animals and Toxins Including Tropical Diseases. 2012;18:198-207.
Crossref - Tempone AG, Andrade Jr HF, Spencer PJ, Lourenco CO, Rogero JR, Nascimento N. Bothrops moojeni venom kills Leishmania spp. with hydrogen peroxide generated by its L-amino acid oxidase. Biochem Biophys Res Commun. 2001;280(3):620-624.
Crossref - Izidoro LFM, Ribeiro MC, Souza GRL. et al. Biochemical and functional characterization of an l-amino acid oxidase isolated from Bothrops pirajai snake venom. Bioorganic Med Chem. 2006;14(20):7034-7043.
Crossref - Okubo BM, Silva ON, Migliolo L, et al. Evaluation of an antimicrobial L-amino acid oxidase and peptide derivatives from Bothropoides mattogrosensis pitviper venom. PLoS One. 2012;7(3):1-10.
Crossref - Izidoro LFM, Sobrinho JC, Mendes MM, et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. Biomed Res Int. 2014;2014(1):196754.
Crossref - Chen LW, Kao PH, Fu YS, Hu WP, Long-Sen C. Bactericidal effect of Naja nigricollis toxin g is related to its membrane-damaging activity. Peptides. 2011;32(8):1755-1763.
Crossref - Chen LW, Kao PH, Fu YS, Lin SR, Chang LS. Membrane-damaging activity of Taiwan cobra cardiotoxin 3 is responsible for its bactericidal activity. Toxicon. 2011;58(1):46-53.
Crossref - Klein RC, Fabres-Klein MH, de Oliveira LL, Feio RN, Malouin F, de Oliveira Barros Ribon A. A C-type lectin from Bothrops jararacussu venom disrupts staphylococcal biofilms. PLoS One. 2015;10(3):1-16.
Crossref - Nunes EdS, de Souza MAA, de Melo Vaz AF, et al. Purification of a lectin with antibacterial activity from Bothrops leucurus snake venom. Comp Biochem Physiol Part B. 2011;159(1):57-63.
Crossref - Joo HS, Otto M. Mechanisms of resistance to antimicrobial peptides in Staphylococci. Biochim Biophys Acta. 2015;1848(11 Pt B):3055-3061.
Crossref - Samy RP, StilesBG, Franco OL, Sethi G, Lim LHK. Animal venoms as antimicrobial agents. Biochem Pharmacol. 2017;134:127-138.
Crossref - Diniz-Sousa R, da S Caldeira CA, Pereira SS, et al. Therapeutic applications of snake venoms: An invaluable potential of new drug candidates. Int J Biol Macromol. 2023;238:124357.
Crossref - Zhao F, Lan X-Q, Du Y, et al. King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates. Zool Res. 2018;39(2):87-96.
Crossref - Almeida JR, Mendes B, Lancellotti M, et al. A novel synthetic peptide inspired on Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom active against multidrug-resistant clinical isolates. Eur J Med Chem. 2018;149:248-256.
Crossref - Power I. An update on analgesics. Br J Anaesth. 2011;107(1):19-24.
Crossref - Kadu AS, Asutkar RD, Chalakh S. A Review on Snake Venom: an Unrevealed Medicine for Human Ailments: Great Scope for Pharmaceutical Research. Int J Res Ayurveda Pharm. 2017;8(2):35-41.
Crossref - Chan YS, Cheung RCF, Xia L, Wong JH, Ng TB, Chan WY. Snake venom toxins: toxicity and medicinal applications. Appl Microbiol Biotechnol. 2016;100(14):6165-6181.
Crossref - Rivera-de-Torre E, Rimbault C, Jenkins TP, et al. Strategies for Heterologous Expression, Synthesis, and Purification of Animal Venom Toxins. Front Bioeng Biotechnol. 2022;9:1-26.
Crossref - Chu XP, Xiong ZG. Acid-sensing ion channels in pathological conditions. In Sodium Calcium Exchange: A Growing Spectrum of Pathophysiological Implications: Proceedings of the 6th International Conference on Sodium Calcium Exchange (pp. 419-431). Boston, MA: Springer US. 2012.
Crossref - Flemming A. Analgesics: Deadly snake venom for pain relief? Nat Rev Drug Discov. 2012;11:906-907.
Crossref - Gomes A. Snake Venom – An Anti-Arthritis Natural Product. Al Ameen J Med Sci. 2010;3(3):176.
- Gomes A, Datta P, Das T, Biswas AK, Gomes A. Anti arthritic and anti inflammatory activity of a cytotoxic protein NN-32 from Indian spectacle cobra (Naja naja) venom in male albino rats. Toxicon. 2014;90:106-110.
Crossref - Nunes FPB, Sampaio SC, Santoro ML, Sousa-e-Silva MCC. Long-lasting anti-inflammatory properties of Crotalus durissus terrificus snake venom in mice. Toxicon. 2007;49(8):1090-1098.
Crossref - Thangam R, Gunasekaran P, Kaveri K, et al. A novel disintegrin protein from Naja naja venom induces cytotoxicity and apoptosis in human cancer cell lines in vitro. Process Biochem. 2012;47(8):1243-1249.
Crossref - Bialves TS, Bastos Jr CLQ, Cordeiro MF, Boyle RT. Snake venom, a potential treatment for melanoma. A systematic review. Int J Biol Macromol. 2023;231:123367.
Crossref - Borkow G, Ovadia M. Selective lysis of virus-infected cells by cobra snake cytotoxins: A sendai virus, human erythrocytes, and cytotoxin model. Biochem Biophys Res Commun. 1999;264:63-68.
Crossref - Ferreira SH. A Bradykinin-Potentiating Factor (Bpf) Present in the Venom of Bothrops Jararaca. Br J Pharmacol Chemother. 1965;24(1):163-169.
Crossref - Petricevich VL, Mendonca RZ. Inhibitory potential of Crotalus durissus terrificus venom on measles virus growth. Toxicon. 2003;42(2):143-153.
Crossref - Neri P, Bracci L, Rustici M, Santucci A. Sequence homology between HIV gp120, rabies virus glycoprotein, and snake venom neurotoxins. Arch Virol. 1990;114(3-4):265-269.
Crossref - Sonnerborg A, Johansson B. The neurotoxin-like sequence of human immunodeficiency virus GP120: A comparison of sequence data from patients with and without neurological symptoms. Virus Genes. 1993;7(1):23-31.
Crossref - da Silva JO, Fernandes RS, Ticli FK, et al. Triterpenoid saponins, new metalloprotease snake venom inhibitors isolated from Pentaclethra macroloba. Toxicon. 2007;50(2):283-291.
Crossref - Fenard D, Lambeau G, Valentin E, Lefebvre JC, Lazdunski M, Doglio A. Secreted phospholipases A2, a new class of HIV inhibitors that block virus entry into host cells. J Clin Invest. 1999;104(5):611-618.
Crossref - Brown AE, Vahey MT, Zhou SYJ, et al. Quantitative relationship of circulating p24 antigen with human immunodeficiency virus (HIV) RNA and specific antibody in hiv-infected subjects receiving antiretroviral therapy. J Infect Dis. 1995;172(4):1091-1094.
Crossref - Zhang YJ, Wang J-H, Lee W-H, et al. Molecular characterization of Trimeresurus stejnegeri venom L-amino acid oxidase with potential anti-HIV activity. Biochem Biophys Res Commun. 2003;309(3):598-604.
Crossref - Nair DG, Fry BG, Alewood P, Kumar PP, Kini RM. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem J. 2007;402(1):93-104.
Crossref - Rádis-Baptista G, Moreno FBMB, Nogueira LDL, et al. Crotacetin, a novel snake venom C-type lectin homolog of convulxin, exhibits an unpredictable antimicrobial activity. Cell biochemistry and Biophysics. 2006;44(3): 412-423.
Crossref - Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657-700.
Crossref - Fenard D, Lambeau G, Maurin T, Lefebvre JC, Doglio A. A peptide derived from bee venom-secreted phospholipase A2 inhibits replication of T-cell tropic HIV-1 strains via interaction with the CXCR4 chemokine receptor. Mol Pharmacol. 2001;60(2):341-347.
Crossref - Villarrubia VG, Costa LA, Diez RA. Secreted phospholipases A₂ (sPLA₂): Friends or foes? Players in antibacterial resistance and human immunodeficiency virus defense? Medicina Clínica (Barcelona). 2004;123(19):749-757.
Crossref - Zack HOM, Oppenheim JJ, Wang JM, Chemokines as molecular targets for therapeutic intervention. J Clin Immunol. 1999;19(5):280-292.
Crossref - Siniavin AE, Streltsova MA, Nikiforova MA, et al. Snake venom phospholipase A2s exhibit strong virucidal activity against SARS-CoV-2 and inhibit the viral spike glycoprotein interaction with ACE2. Cell Mol Life Sci. 2021;78(23):7777-7794.
Crossref - El-Aziz TMA, Soares AG, Stockand JD. Snake venoms in drug discovery: Valuable therapeutic tools for life saving. Toxins (Basel). 2019;11(10):1-25.
Crossref - Cherifi F, Laraba-Djebari F. Bioactive Molecules Derived from Snake Venoms with Therapeutic Potential for the Treatment of Thrombo-Cardiovascular Disorders Associated with COVID-19. Protein J. 2021;40(6):799-841.
Crossref - de Castro Figueiredo BK, Cologna CT, Fornari-Baldo EC, et al. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front Pharmacol. 2020;11.
Crossref - Rao WQ, Kalogeropoulos K, Allentoft M, et al. The rise of genomics in snake venom research: recent advances and future perspectives. Gigascience. 2022;11:giac024.
Crossref - Post Y, Puschhof J, Beumer J, et al. Snake Venom Gland Organoids. Cell. 2020;180(2):233-247.e21.
Crossref - Zhang L, Falla TJ. Cosmeceuticals and peptides. Clin Dermatol. 2009;27(5):485-494.
Crossref - Derakhshani A, Silvestris N, Hajiasgharzadeh K, et al. Expression and characterization of a novel recombinant cytotoxin II from Naja naja oxiana venom: A potential treatment for breast cancer. Int J Biol Macromol. 2020;162:1283-1292.
Crossref - Romero-Giraldo LE, Pulido S, Berrio MA, et al. Heterologous Expression and Immunogenic Potential of the Most Abundant Phospholipase A2 from Coral Snake Micrurus dumerilii to Develop Antivenoms. Toxins (Basel). 2022;14(12):825.
Crossref - Chakrabarty D, Chanda C. Snake Venom Disintegrins. Snake Venoms. 2017:437-449.
Crossref - Trifiro G, Gini R, Barone-Adesi F, et al. The Role of European Healthcare Databases for Post-Marketing Drug Effectiveness, Safety and Value Evaluation: Where Does Italy Stand? Drug Saf. 2019;42(2):347-363.
Crossref - Ferrajolo C, Coloma PM, Verhamme KMC, et al. Signal Detection of Potentially Drug-Induced Acute Liver Injury in Children Using a Multi-Country Healthcare Database Network. Drug Saf. 2014;37(2):99-108.
Crossref - Williams DJ, Gutierrez J-M, Calvete JJ, et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J Proteomics. 2011;74(9):1735-1767.
Crossref - Parveen S, Misra R, Sahoo SK. Nanoparticles: A boon to drug delivery, therapeutics, diagnostics and imaging. Nanomedicine Nanotechnology Biol Med. 2012;8(2):147-166.
Crossref - Biswas A, Gomes A, Sengupta J, et al. Nanoparticle-conjugated animal venom-toxins and their possible therapeutic potential. J Venom Res. 2012;3:15-21.
- Attarde SS, Pandit SV. Anticancer potential of nanogold conjugated toxin GNP-NN-32 from Naja naja venom. J Venom Anim Toxins Incl Trop Dis. 2020;26:1-12.
Crossref - Bhowmik T, Saha PP, Dasgupta A, Gomes A. Antileukemic potential of PEGylated gold nanoparticle conjugated with protein toxin (NKCT1) isolated from Indian cobra (Naja kaouthia) venom. Cancer Nano. 2013;4(1-3):39-55.
Crossref - Mohammadur Dounighi N, Mehrabi M, Avadi MR, Zolfagharian H, Rezayat M. Preparation, characterization and stability investigation of chitosan nanoparticles loaded with the Echis carinatus snake venom as a novel delivery system. Archives of Razi Institute. 2015;70(4):269-277.
Crossref - Badr G, Sayed D, Maximous D, Mohamed AO, Gul M. Increased susceptibility to apoptosis and growth arrest of human breast cancer cells treated by a snake venom-loaded silica nanoparticles. Cell Physiol Biochem. 2014;34(5):1640-1651.
Crossref - Gouveia DM, Cogo JC, Kido HW, et al. Physical-chemical characterization of carbon nanotubes-hydroxyapatite nanocomposites with fibrin sealant derived from snake venom. Toxicon. 2020;177(Suppl 1):S56.
Crossref - Mladic M, Slagboom J, Kool J, Vonk FJ , van Wezel GP, Richardson MK. Detection and identification of antibacterial proteins in snake venoms using at-line nanofractionation coupled to LC-MS. Toxicon. 2018;155:66-74.
Crossref - Lu Q, Navdaev A, Clemetson JM, Clemetson KJ. Snake venom C-type lectins interacting with platelet receptors. Structure-function relationships and effects on haemostasis. Toxicon. 2005;45(8):1089-1098.
Crossref - Stabeli RG, Marcussi S, Carlos GB, et al. Platelet aggregation and antibacterial effects of an L-amino acid oxidase purified from Bothrops alternatus snake venom. Bioorganic Med Chem. 2004;12(11):2881-2886.
Crossref - Sun MZ, Gao C, Tian Y, Chen D, Greenaway FT, Liu S. Biochemical, functional and structural characterization of Akbu-LAAO: A novel snake venom l-amino acid oxidase from Agkistrodon blomhoffii ussurensis. Biochimie. 2010;92(4):343-349.
Crossref - Tonismagi K, Samel M, Trummal K, et al. L-Amino acid oxidase from Vipera lebetina venom: Isolation, characterization, effects on platelets and bacteria. Toxicon. 2006;48(2):227-237.
Crossref - Toyama MH, de O Toyama D, Passero LFD, et al. Isolation of a new L-amino acid oxidase from Crotalus durissus cascavella venom. Toxicon. 2006;47(1):47-57.
Crossref - Findrik Z, Geueke B, Hummel W, Vasic-Racki D. Modelling of L-DOPA enzymatic oxidation catalyzed by L-amino acid oxidases from Crotalus adamanteus and Rhodococcus opacus. Biochem Eng J. 2006;27(3):275-286.
Crossref - Phua CS, Vejayan J, Ambu S, Ponnudurai G, Gorajana A. Purification and antibacterial activities of an L-amino acid oxidase from king cobra (Ophiophagus hannah) venom. J Venom Anim Toxins Incl Trop Dis. 2012;18(2):198-207.
Crossref - San TM, Vejayan J, Shanmugan K, Ibrahim H. Screening antimicrobial activity of venoms from snakes commonly found in Malaysia. J Appl Sci. 2010;10(19):2328-2332.
Crossref - Lomonte B, Angulo Y, Sasa M, Gutierrez JM. The Phospholipase A2 Homologues of Snake Venoms: Biological Activities and Their Possible Adaptive Roles. Protein Pept Lett. 2009;16(8):860-876.
Crossref - Samy RP, Gopalakrishnakone P, Thwin MM, et al. Antibacterial activity of snake, scorpion and bee venoms: A comparison with purified venom phospholipase A2 enzymes. J Appl Microbiol. 2007;102(3):650-659.
Crossref - Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50 Suppl (Suppl):237-242.
Crossref - Abdullahi ZU, Musa SS, Abu-Odah H, Ahmed A, Lawan AA, Bello UM. Bactericidal effects of snake venom phospholipases A2: a systematic review and analysis of minimum inhibitory concentration. Physiologia, 2023; 3(1):30-42.
Crossref - Fung SY, Lee ML, Tan NH. Molecular mechanism of cell death induced by king cobra (Ophiophagus hannah) venom l-amino acid oxidase. Toxicon, 2015;96:38-45.
Crossref - Zouari-Kessentini R, Srairi-Abid N, Bazaa A, El Ayeb M, Luis J, Marrakchi N. Antitumoral potential of Tunisian snake venoms secreted phospholipases A2. Biomed Res. Int. 2013;2013(1):391389.
Crossref - Feng L, Gao R, Gopalakrishnakone P. Isolation and characterization of a hyaluronidase from the venom of Chinese red scorpion Buthus martensi. Comp Biochem Physiol C Toxicol Pharmacol. 2008;148(3):250-257.
Crossref - Marcinkiewicz C, Lobb RR, Marcinkiewicz MM, et al. Isolation and characterization of EMS16, a C-lectin type protein from Echis multisquamatus venom, a potent and selective inhibitor of the a2b1 integrin. Biochemistry. 2000;39(32):9859-9867.
Crossref - Markland FS, Swenson S. Snake venom metalloproteinases. Toxicon. 2013;62:3-18.
Crossref - Staniszewska I, Walsh EM, Rothman VL, et al. Effect of VP12 and viperistatin on inhibition of collagen receptors-dependent melanoma metastasis. Cancer Biol Ther. 2009;8(15):1507-1516.
Crossref - de Carvalho DD, Schmitmeier S, Novello JC, Markland FS. Effect of BJcuL (a lectin from the venom of the snake Bothrops jararacussu) on adhesion and growth of tumor and endothelial cells. Toxicon. 2001;39(10):1471-1476.
Crossref - Marcinkiewicz C, Weinreb PH, Calvete JJ, et al. Obtustatin: A potent selective inhibitor of a1b1 integrin in vitro and angiogenesis in vivo. Cancer Res. 2003;63(9):2020-2023.
- Olfa KZ, Jose L, Salma D, et al. Lebestatin, a disintegrin from Macrovipera venom, inhibits integrin-mediated cell adhesion, migration and angiogenesis. Lab Investig. 2005;85(12):1507-1516.
Crossref - Yang RS, Tang CH, Chuang WJ, et al. Inhibition of tumor formation by snake venom disintegrin. Toxicon. 2005;45(5):661-669.
Crossref - Sahni A, Simpson-haidaris PJ, Sahni SK, Vaday GG, Francis CW. Fibrinogen synthesized by cancer cells augments the proliferative effect of fibroblast growth factor-2 (FGF-2). J Thromb Haemost. 2008;6(1):176-183.
Crossref - Son DJ, Park MH, Chae SJ, et al. Inhibitory effect of snake venom toxin from Vipera lebetina turanica on hormone-refractory human prostate cancer cell growth: Induction of apoptosis through inactivation of nuclear factor kB. Mol Cancer Ther. 2007;6(2):675-683.
Crossref - Dong SK, Jang YJ, Jeon OH, Kim DS. Saxatilin, a snake venom disintegrin, suppresses TNF-a-induced ovarian cancer cell invasion. J Biochem Mol Biol. 2007;40(2):290-294.
Crossref - Sarray S, Delamarre E, Marvaldi J, El Ayeb M, Marrakchi N, Luis J. Lebectin and lebecetin, two C-type lectins from snake venom, inhibit a5b1 and av-containing integrins. Matrix Biol. 2007;26(4):306-313.
Crossref - Tian J, Paquette-Straub C, Sage EH, et al. Inhibition of melanoma cell motility by the snake venom disintegrin eristostatin. Toxicon. 2007;49(7):899-908.
Crossref - Premzl A, Kovacic L, Halassy B, Krizaj I. Generation of ammodytoxin-anti-cathepsin B immuno-conjugate as a model for delivery of secretory phospholipase A2 into cancer cells. Toxicon. 2008;51(5):754-764.
Crossref - Galan JA, Sanchez EE, Rodriguez-Acosta A, et al. Inhibition of lung tumor colonization and cell migration with the disintegrin crotatroxin 2 isolated from the venom of Crotalus atrox. Toxicon. 2008;51(7):1186-1196.
Crossref - Shin VY, Jin HC, Ng EKO. et al. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induce cyclooxygenase-2 activity in human gastric cancer cells: Involvement of nicotinic acetylcholine receptor (nAChR) and b-adrenergic receptor signaling pathways. Toxicol Appl Pharmacol. 2008;233(2):254-261.
Crossref - Kessentini-Zouari R, Jebali J, Taboubi S, et al. CC-PLA2-1 and CC-PLA2-2, two Cerastes cerastes venom-derived phospholipases A2, inhibit angiogenesis both in vitro and in vivo. Lab Investig. 2010;90(4):510-519.
Crossref - Jang SH, Ryu PD, Lee SY. Dendrotoxin-k suppresses tumor growth induced by human lung adenocarcinoma A549 cells in nude mice. J Vet Sci. 2011;12(1):35-40.
Crossref - Lucena S, Sanchez EE, Perez JC. Anti-metastatic activity of the recombinant disintegrin, r-mojastin 1, from the Mohave rattlesnake. Toxicon. 2011;57(5):794-802.
Crossref - Higuchi DA, Almeida MC, Barros CC, et al. Leucurogin, a new recombinant disintegrin cloned from Bothrops leucurus (white-tailed-jararaca) with potent activity upon platelet aggregation and tumor growth. Toxicon. 2011;58(1):123-129.
Crossref - Naumann GB, Silva LF, Silva L, et al. Cytotoxicity and inhibition of platelet aggregation caused by an l-amino acid oxidase from Bothrops leucurus venom. Biochim Biophys Acta. 2011;1810(7):683-694.
Crossref - Minea R, Helchowski C, Rubino B, Brodmann K, Swenson S, Markland Jr F. Development of a chimeric recombinant disintegrin as a cost-effective anti-cancer agent with promising translational potential. Toxicon. 2012;59(4):472-486.
Crossref - Yalcin HT, Ozen MO, Gocmen B, Nalbantsoy A. Effect of Ottoman Viper (Montivipera xanthina (Gray, 1849)) Venom on Various Cancer Cells and on Microorganisms. Cytotechnology. 2014;66(1):87-94.
Crossref - Lucena S, Castro R, Lunding C, et al. Inhibition of pancreatic tumoral cells by snake venom disintegrins. Toxicon. 2015;93:136-143.
Crossref - Saviola AJ, Burns PD, Mukherjee AK, Mackessy SP. The disintegrin tzabcanin inhibits adhesion and migration in melanoma and lung cancer cells. Int J Biol Macromol. 2016;88:457-464.
Crossref - Kerkkamp H, Bagowski C, Kool J, van Soolingen B, Vonk FJ, Vlecken D. Whole snake venoms: Cytotoxic, anti-metastatic and antiangiogenic properties. Toxicon. 2018;150:39-49.
Crossref - Valente RH, Nicolau CA, Perales J, Neves-Ferreira AGdC. Snake Venom Proteopeptidomics: What Lies Behind the Curtain. In: Gopalakrishnakone, P., Calvete, J. (eds) Venom Genomics and Proteomics. Springer, Dordrecht. 2014.
Crossref - Millgard J, Hagg A, Sarabi M, Lind L. Captopril, but not nifedipine, improves endothelium-dependent vasodilation in hypertensive patients. J Hum Hypertens. 1998;12:511-516.
Crossref - Waheed H, Moin SF, Choudhary MI. Snake Venom: From Deadly Toxins to Life-saving Therapeutics. Curr Med Chem. 2017;24(17):1874-1891.
Crossref - Kuo YJ, Chung CH, Huang TF. From discovery of snake venom disintegrins to a safer therapeutic antithrombotic agent. Toxins (Basel). 2019;11(7):1-11.
Crossref - Scarborough RM, Rose JW, Hsu MA, et al. Barbourin: A GPIIb-IIIa-Specific integrin antagonist from the venom of Sistrurus M. Barbouri. J Biol Chem. 1991;266(15):9359-9362.
Crossref - Lang SH, Manning N, Armstrong N, et al. Treatment with tirofiban for acute coronary syndrome (ACS): A systematic review and network analysis. Curr Med Res Opin. 2012;28(3):351-370.
Crossref - Lazarovici P, Marcinkiewicz C, Lelkes PI. From snake venom’s disintegrins and C-type lectins to anti-platelet drugs. Toxins (Basel). 2019;11(5):1-15.
Crossref - Topol, E. J., Byzova, T. V., & Plow, E. F. (1999). Platelet GPIIb-IIIa blockers. The Lancet, 353 (9148):227-231.
Crossref - Vu TT, Stafford AR, Leslie BA, Kim PY, Fredenburgh JC, Weitz JI. Batroxobin binds fibrin with higher affinity and promotes clot expansion to a greater extent than thrombin. J Biol Chem. 2013;288(23):16862-16871.
Crossref - Sajevic T, Leonardi A, Krizaj I. Haemostatically active proteins in snake venoms. Toxicon. 2011;57(5):627-645.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.