ISSN: 0973-7510
E-ISSN: 2581-690X
Lignocellulose is a core component of plant biomass and the most abundant carbohydrate polymer in nature. It is cheap and renewable and has several potential applications; however, it remains underutilized because of its recalcitrance to degradation. Cellulolytic microbes have been found in the gut of herbivorous insects, such as grasshoppers. This study aimed to isolate lignocellulolytic bacteria from the gut of grasshoppers (Oxya chinensis) and determine their diversity and potential biomass-degrading activity. A total of 27 culturable isolates were obtained from the grasshopper foregut, midgut, and hindgut. The bacteria hydrolyzed cellulose and lignin, as indicated by a cellulolytic index of 0.12–1.23 and ligninolytic index of 0.1–1.47. Five potential cellulolytic bacterial isolates were selected. Based on 16S rRNA sequencing, the isolates were identified as Bacillus wiedmannii (foregut), Bacillus marcorestinctum, Bacillus halotolerans (midgut), Paenibacillus zanthoxyli, and Bacillus hominis (hindgut). The highest specific cellulolytic activity (0.0068 U/mg) was detected in B. wiedmannii (OCF2), which could be exploited as a potential source of cellulases.
Bacillus, Cellulolytic Bacteria, Enzymatic Activity, Lignocellulosic Biomass
Lignocellulosic biomass is the most abundant carbohydrate polymer in nature and one of the cheapest renewable resources. Lignocellulose is a fundamental component of plants and is extensively exploited by industries such as pharmaceutical, biofuels, and food production.1,2 Lignocellulose is comprised of cellulose (40%–60%), hemicellulose (20%–40%), and lignin (10%–25%).3 Cellulose constitutes the primary constituent of the cellular wall in plants. It is water-insoluble, with a fibrous, stringent, crystalline form, and a renewable source of energy in the biosphere.4,5 Hemicellulose consists primarily of xylan, which comprises a main chain of β-1,4-linked xylopyranose residues.3,6 Lignin, an aromatic biopolymer, is widely prevalent in nature and constitutes approximately 30% of the mass found in the secondary wall of plants.7
Lignocellulosic biomass can be hydrolyzed either chemically or enzymatically. The existence of symbiotic bacteria in the digestive tract allows invertebrates and herbivorous animals to digest lignocellulose. Lignocellulolytic enzymes in insects can be derived from gut bacterial symbionts, fungi or the host organism itself.8 The gut microbes has the ability to break down lignocellulose to release glucose and other fermentable sugars from complex polysaccharides in the plant cell walls.9 Cellulases are extracellular enzymes capable of converting cellulose to simple sugars10 by breaking 1,4-β-glycosidic bonds in cellulose, cellodextrin, cellobiose, and other cellulose derivatives.11,12
The use of bacteria as a source of cellulose-degrading enzymes has several advantages, namely low production costs, rapidity, simplicity, consistent output, and easy control.13 Insects possess the ability to produce digestive enzymes through the assistance of symbiotic bacteria residing within their bodies. This mechanism facilitates the efficient breakdown of food and acquisition of energy necessary for their personal growth and developmental processes.14 Microbes found in the gut of host organisms thrive on lignocellulosic biomass as their major feed.15 Numerous phytophagous insects, including termites, beetles, and wasps, have highly effective mechanisms for converting lignocellulose.16 Some groups of arthropoda, such as millipede, have been reported unable to degrade all lignocellulose components. The cellulolytic enzyme was not detected in the insect has amylase activity.17
The gut microbiota of grasshoppers is thought to content various type microbes, helping these animals to extract nutrients from plant material.18 Several bacterial phyla have been documented in the gastrointestinal tract of grasshoppers, including Firmicutes, Proteobacteria, and Actinobacteria.19 Bacteria are believed to possess lignocellulolytic activities under both aerobic and anaerobic conditions.20 Some of the anaerobic bacteria include Acetivibrio, Bacteroides, and Clostridium; whereas the aerobic ones include Bacillus sp., Cellulomonas sp., and Pseudomonas.21 Microbes found in the insect gut, including Pseudomonas, Bacillus, Enterobacter, and Paenibacillus, are known to produce lignocellulosic enzymes.22 Complex interactions exist between gut microbes and their insect hosts to the benefit of both parties.8 Cellulolytic bacterial isolates were previously reported in the gut of the grasshoppers Oxya chinensis and Oxya velox.23,24 The present study aimed to explore biomass degrading activity of bacteria isolated from the gut of O. chinensis.
Sample collection
Four grasshoppers were collected with a net in the agricultural area of Medan Selayang sub-district, Indonesia. The collected grasshoppers were at the 4th instar or higher stage.17 The grasshoppers were placed in containers, where they had no access to food until dissection and identification in the laboratory.3
Isolation of bacteria from the grasshopper gut
Prior to gut dissection, grasshoppers were surface-sterilized with 70% ethanol and kept on ice for 15 min. The entire intestinal tract was removed and divided into three parts: foregut, midgut, and hindgut.3 The contents of each part were suspended in water and serially diluted to 10-6. At each dilution, 0.1 mL was taken and inoculated in a Petri dish containing nutrient agar medium.25 The dishes were incubated at 37°C for 24 h, and the number of bacterial colonies was counted. The isolates were characterized both macroscopically and microscopically.26
Screening for cellulolytic bacteria
Screening for cellulolytic activity of isolates was done following previously described method.27 Briefly, they were grown in carboxymethyl cellulose (CMC) agar medium containing 1.0 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 0.5 g/L NaCl, 0.01 g/L FeSO4·7H2O, 0.01 g/L MnSO4·H2O, 0.3 g/L NH4NO3, 10.0 g/L CMC, and 12.0 g/L agar. Each bacterial isolate was inoculated at the center of the plate, and the plate was incubated at ± 32°C for 3 days. Subsequently, the dish was flooded with Congo red (0.1% w/v), left covered for 15 min, rinsed with 1 M NaCl, and allowed to stand for 15-20 min.28 All experiments were performed in triplicate. The clear zone formed around the colony was measured with a caliper and was used to calculate the cellulolytic index.
Cellulolytic index=((Diameter of clear zone-Diameter of bacterial colony))/(Diameter of bacterial colony)
Screening for ligninolytic bacteria
Ligninolytic activity was screened as described previously.29 Briefly, the isolates were grown on medium containing 5 g/L glucose, 5.0 g/L yeast extract, 2 g/L KH2PO4, 2 g/L Na2HPO4, 0.05 g/L MgSO4·7H2O, 0.01 g/L CaCl2·2H2O, 0.01 g/L CuSO4·5H2O, and 15.0 g/L agar. Following sterilization, methylene blue to a final concentration of 1 g/L was added to the medium. Each isolate was inoculated on the agar plate, and the plate was incubated at 32°C for a week. The clear zone formed around the colony was measured with a caliper and was used to calculate the ligninolytic index, based on the same formula used for the cellulolytic index.
Xylanolytic activity was screening as described previously.6 Each isolate was spot-inoculated on xylan agar medium containing 0.5% (w/v) birch wood xylan, 0.5% (w/v) yeast extract, 0.5% (w/v) peptone, 0.02% (w/v) MgSO4.7H2O, 0.1% (w/v), K2HPO4, and 2.0% (w/v) agar. Each bacterial isolate was inoculated in the center of the plate, and the plate was incubated at ± 32°C for 3 days. Subsequently, the dishes were flooded with 0.4% Congo red and, after 10 min, rinsed with 1 M NaCl.30 The clear zone surrounding the colony was measured with a caliper and was used to calculate the xylanolytic index, based on the same formula used for the cellulolytic index.
Cellulase production
Enzyme production was assessed by inoculating selected bacterial isolates into CMC liquid medium containing basal salt medium without agar (1.0 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 0.5 g/L NaCl, 0.01 g/L FeSO4·7H2O, 0.01 g/L MnSO4·H2O, 0.3 g/L NH4NO3), followed by incubation at 37°C with aeration at 120 rpm for 8 days.31 Subsequently, 5 mL of culture medium suspension was centrifuged for 10 min at 6,000 rpm and 4°C. The supernatant served as a crude extract for quantifying enzyme activity and dissolved protein levels.28
Cellulase activity assay
Cellulolytic activity was measured following the modified method of Miller (1959).32 Briefly, 1 mL crude extract enzyme was pipetted to 1 mL of 1% 3,5-dinitrosalicylic acid (CMC), and incubated for 60 min at 37°C. Following the incubation 1 mL of 20 mM phosphate buffer (pH 7) and 2 mL DNS were added to the mixture. The sample mixture was vortexed and placed in a water bath at 100°C for 10 min. The blank contained 1 mL distilled water instead of crude extract. The absorbance was recorded at 540 nm.15 One unit of enzymatic activity was defined as the amount of enzyme that produced 1 µmol of glucose per mL per min.33 A standard curve with glucose was plotted to calculate cellulase activity in the sample. Protein content was measured following Lowry’s method at 595 nm and bovine serum albumin was used as a standard.5 The calculated enzyme level was then used to determine the specific activity of the enzyme based on the following formula:11
Specific Activity (U/mg)=(Enzyme Activity)/(Protein Content)
Molecular identification of potential bacteria
Five cellulolytic isolates with higher cellulolytic indexes were characterized based on 16S rRNA sequences. DNA was isolated by freezing and thawing. One inoculation loop of bacterial suspension was transferred to Eppendorf tube with 100 ml of sterile double distilled water. The tube was chilled (-10°C) for 10 minutes and thawed for 10 minutes at 90°C. The step was repeated five times.34 The suspension was centrifuged at 10,000 x g for 10 min. The supernatant was kept at 4°C. The isolated DNA was used as a template for the amplification of the 16S rRNA gene using the bacterial universal primers 27F (5′-AGAGTTTGATCCTGGTCCAG-3′) and 1492R (5′-CTACGGGCTACCTTGTTACGA-3′).35 The reaction was prepared in a total volume of 25 µL with the following program: pre-denaturation at 94°C for 2 min; 40 cycles of denaturation at 92°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 1 min; and post-elongation at 72°C for 5 min.34 The PCR results were run along with a 1-kb DNA marker on a 1% agarose gel at 80 V and 400 mA for 60 min for monitoring the results. The PCR product was sent to Macrogen Singapore for sequencing.
Bioinformatics analysis
The nucleotide sequence of the 16S ribosomal DNA (rDNA) gene from the five selected isolates was aligned and compared with 16S ribosomal RNA (rRNA) data available in GenBank using the BLASTn tool.36 Analysis of bacterial kinship based on phylogenetic trees was performed in MEGA XI software with 1,000 bootstrap repetitions.37,38
Bacterial isolates from the grasshopper gut
The diversity and role of bacteria resides on the digestive system of animals has been studied from different viewpoints, and different approaches have been done to isolate types of microorganism. Koubova et al.39 isolated actinobacteria, bacteria and fungi from millipede Telodeinopus aoutii gut. Willis et al. 40 isolated bacteria from the foregut and hindgut of the Carolina wasp Dissosteira carolina. Here, bacteria were successfully isolated from all three parts of the grasshopper gut, yielding a total of 27 culturable isolates: 9 from the foregut, 10 from the midgut, and 8 from the hindgut. Previously, Shil et al.23 recovered and identified 15 bacterial isolates from the gut of O. velox. Abdullah et al. 41 successfully isolated 80 bacterial types from Valanga nigricornis. Wang et al.42 isolated bacteria from three different grasshoppers: Aiolopus tamulus (31 isolates), Oedaleus decorus asiaticus (32 isolates), and Shirakiacris shirakii (30 isolates).
Based on the macroscopic characterization reported in Table 1, the bacterial colonies presented mostly an irregular shape, milky white color, complete margin, and raised elevation. Gram staining revealed 22 gram-positive bacteria and 5 gram-negative bacteria. The different shape and growth of each isolate indicated that they originated from different types of bacteria.43 The highest number of isolates was obtained from the midgut and the smallest from the hindgut. According to Arfah et al.,11 many digestive processes, such as the absorption of carbohydrates, proteins, and lipids, occur in the midgut of grasshoppers; however, the distribution of bacteria in the foregut and midgut differs between that of O. chinensis and Oedaleus infernalis.44
Table (1):
Morphological characteristics of bacterial isolates from the grasshopper gut
| Isolate code | Bacterial colonies | Cell Shape/ Gram stain | |||
|---|---|---|---|---|---|
| Shape | Color | Margin | Elevation | ||
| Foregut | |||||
| OCF1 | Irregular | Pale white | Undulate | Raised | Bacilli / (−) |
| OCF2 | Circular | White | Entire | Flat | Bacilli / (+) |
| OCF3 | Circular | Pale white | Entire | Umbonate | Bacilli / (−) |
| OCF4 | Irregular | Pale white | Undulate | Raised | Bacilli / (+) |
| OCF5 | Irregular | Pale white | Undulate | Raised | Bacilli / (+) |
| OCF6 | Irregular | White transparent | Undulate | Umbonate | Bacilli / (+) |
| OCF7 | Irregular | Red | Entire | Raised | Bacilli / (+) |
| OCF8 | Irregular | Red | Entire | Raised | Cocci / (−) |
| OCF9 | Irregular | Pale white | Entire | Raised | Bacilli / (+) |
| Midgut | |||||
| OCM1 | Irregular | Pale white | Undulate | Raised | Bacilli / (+) |
| OCM2 | Circular | Pale white | Entire | Raised | Cocci / (+) |
| OCM3 | Irregular | White | Undulate | Raised | Cocci / (−) |
| OCM4 | Circular | Yellow white | Entire | Convex | Bacilli / (+) |
| OCM5 | Circular | Red | Entire | Convex | Cocci / (−) |
| OCM6 | Irregular | White transparent | Entire | Umbonate | Bacilli / (+) |
| OCM7 | Irregular | Pale white | Undulate | Umbonate | Bacilli / (+) |
| OCM8 | Circular | Yellow white | Entire | Raised | Bacilli / (+) |
| OCM9 | Irregular | Pale white | Entire | Convex | Bacilli / (+) |
| OCM10 | Circular | Pale white | Entire | Convex | Bacilli / (+) |
| Hindgut | |||||
| OCH1 | Irregular | White | Entire | Raised | Bacilli / (+) |
| OCH2 | Circular | Pale white | Entire | Convex | Bacilli / (+) |
| OCH3 | Irregular | White | Entire | Raised | Bacilli / (+) |
| OCH4 | Irregular | Pale white | Entire | Raised | Bacilli / (+) |
| OCH5 | Circular | White transparent | Entire | Umbonate | Bacilli / (+) |
| OCH6 | Irregular | Pale white | Undulate | Umbonate | Bacilli / (+) |
| OCH7 | Irregular | White | Entire | Umbonate | Bacilli / (+) |
| OCH8 | Circular | Yellow white | Entire | Raised | Bacilli / (+) |
Gram stain: +, gram-positive; −, gram-negative.
Lignocellulolytic activity of bacterial isolates
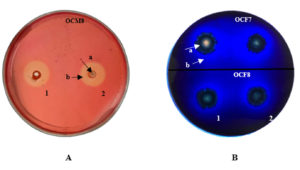
Clear zones were visible around inoculation sites on CMC agar plates flooded with 1% Congo red (Figure 1A) and methylene blue-containing lignin agar plates (Figure 1B). Saini et al.45 stated that the clear zone around a colony indicated extracellular cellulase production by bacteria. Congo red at 1% enables the detection of clear zones hydrolyzed by cellulase.46 Xylanolytic activity was not visible in xylan agar, as no clear zone appeared around the bacterial inocula after flooding the plate with 0.4% Congo red. This finding suggests that the O. chinensis gut bacteria probably cannot utilize the carbon in xylan agar. Alternatively, the screening method employed in this study6,30 may not be suitable for detecting xylanase activity in bacterial samples. Indeed, while xylanase production in fungi is well documented,3 little is known about such enzymes in the insect gut.
Figure 1. Presence of (A) cellulolytic activity in carboxymethyl cellulose agar and (B) lignocellulolytic activity in lignin agar. 1: first repetition; 2: second repetition; a: bacterial colonies; b: clear zone.
The formation of clear zones on methylene blue agar plates indicated that the bacterial colony produced enzymes capable of degrading lignin.43 Dineshkumar et al.46 suggested the use of methylene blue to quantify ligninolytic activity. The decolorization of methylene blue is due to the secretion of extracellular ligninolytic enzymes.47 Isolates that can form a clear zone with a diameter twice that of the colony are considered potential producers of ligninolytic enzymes.48
The enzymatic activity of the different bacterial isolates was calculated based on measuring the diameter of the clear zones around the colonies (Table 2) and the values were the average of three measurements. Bacterial isolates possessed cellulolytic (100%) and ligninolytic (55.5%) activity, but no xylanolytic (0%) activity, suggesting a varying ability to hydrolyze the carbon source. The lignocellulolytic index is strongly influenced by the ability to produce the corresponding enzymes.
Table (2):
Lignocellulolytic index of bacterial isolates from the grasshopper gut
Isolate code |
Cellulolytic index |
Ligninolytic index |
Xylanolytic index |
|---|---|---|---|
OCF1 |
0.45 |
0 |
0 |
OCF2 |
0.86 |
1.04 |
0 |
OCF3 |
0.30 |
1.31 |
0 |
OCF4 |
0.71 |
0 |
0 |
OCF5 |
0.13 |
0.99 |
0 |
OCF6 |
0.29 |
0.71 |
0 |
OCF7 |
0.61 |
1.47 |
0 |
OCF8 |
0.32 |
0.32 |
0 |
OCF9 |
0.37 |
0 |
0 |
OCM1 |
0.41 |
0.64 |
0 |
OCM2 |
0.41 |
0 |
0 |
OCM3 |
0.12 |
1.02 |
0 |
OCM4 |
1.07 |
0 |
0 |
OCM5 |
0.12 |
0.65 |
0 |
OCM6 |
0.50 |
0 |
0 |
OCM7 |
0.20 |
0.4 |
0 |
OCM8 |
1.20 |
1.22 |
0 |
OCM9 |
0.60 |
0 |
0 |
OCM10 |
0.50 |
0.54 |
0 |
OCH1 |
0.37 |
0 |
0 |
OCH2 |
0.42 |
0 |
0 |
OCH3 |
0.53 |
0 |
0 |
OCH4 |
1.06 |
0.1 |
0 |
OCH5 |
0.99 |
0.16 |
0 |
OCH6 |
0.34 |
0.17 |
0 |
OCH7 |
1.23 |
0 |
0 |
OCH8 |
0.91 |
0 |
0 |
The highest cellulolytic index was found in isolates OCH7 (1.23), OCM8 (1.20), and OCM4 (1.07); the highest ligninolytic index was detected in isolates OCF7 (1.47), OCF3 (1.31), and OCM8 (1.22). Notably, the xylanolytic index was zero in all isolates (Table 2). Hence, the bacteria found in the gut of O. chinensis might be unable to use xylan as a carbon source but can readily utilize cellulose and lignin.
Jimenez and Hernandez49 reported that fungi showed greater capability for degrading lignocellulosic materials than bacteria in the gut of wood-feeding Coleoptera, with the genera Trichoderma, Bionectria, and Trametes showing positive results in all the assays performed. Abdullah et al.41 reported a cellulolytic index of 0.9-1.7 for bacterial isolates from the grasshopper V. nigricornis. Ferbiyanto et al.19 isolated bacteria from the gut of the termite Macrotermes gilvus and reported a cellulolytic index of 0.75–2.5. Simol et al.21 reported 10 potential ligninolytic microbes from the gut of the termite Coptotermes curvignathus. Recent work by Koubova et al.39 found that 30% of isolated bacteria and fungi from the gut of millipede T. aoutii exhibited cellulase activity in vitro. They also identified high activity of cellulase from Actinobacteria Streptomyces and Kitasatospora.
Cellulase activity of bacterial isolates
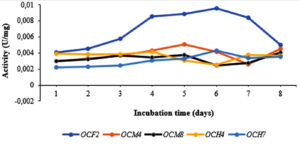
Bacterial isolates with the highest cellulolytic index on plates were incubated in liquid CMC medium for 8 days. Subsequently, cellulase activity, protein level, and specific cellulase activity were measured. As shown in Figure 2, in general, the highest specific cellulase activity was detected on day 5, particularly for OCF2 (0.0088 U/mg) and OCM4 (0.0050 U/mg); the values for OCM8, OCH4, and OCH7 clustered around 0.0037 U/mg, 0.0030 U/mg, and 0.0032 U/mg, respectively. The specific cellulase activity increased in some cases until day 6 and decreased significantly thereafter. The protein level ranged from 88.829 mg/mL to 92.4 mg/mL. Grass-consuming grasshoppers and the wood-consuming wood borer possess higher gut cellulase activities than leaf-consuming silkworms.50 Specifically, cellulase activity in the gut fluid of O. velox was reported as 0.759 ± 0.005 U/mg,3 whereas that of Tribolium castaneum larvae was 0.016 U/mg.51 Sreena et al.52 isolated five strains of cellulolytic bacteria from two termite species and found that the highest endoglucanase activity was that of Bacillus cereus (5.06 U/mg) while the lowest was 2.98 U/mg.
Molecular identification of bacterial isolates
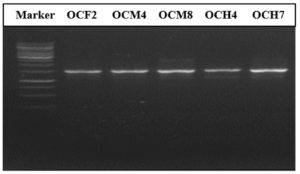
Molecular identification of the bacterial isolates was performed by sequencing their 16S rRNA gene using universal primers. Based on the results in Figure 3, the amplified DNA was 1,500 bp in size. The 16S rRNA PCR products of five potential cellulolytic microbes were purified, sequenced, and aligned for comparison by BLASTn against the NCBI database (Table 3).
Figure 3. PCR results showing amplification of the 16S rRNA gene. Row 1 corresponds to the 1-kb DNA marker. Rows 2–5 correspond to bacterial isolates from the gut of O. chinensis
Table (3):
BLASTn of the 16S rRNA gene from five bacterial isolates
Isolate code |
Closely related bacteria |
GenBank accession no. |
Identity (%) |
|---|---|---|---|
OCF2 |
Bacillus wiedmannii strain FSL W8-0169 |
NR_152692.1 |
93.46 |
OCM4 |
Bacillus marcorestinctum strain LQQ |
NR_117414.1 |
94.03 |
OCM8 |
Bacillus halotolerans strain CR-119 |
NR_115283.1 |
86.15 |
OCH4 |
Paenibacillus zanthoxyli strain JH29 |
NR_043876.1 |
97.37 |
OCH7 |
Bacillus hominis strain BML-BC059 |
NR_175557.1 |
95.79 |
Alignment revealed that the cellulolytic bacteria belonged to two genera: Bacillus and Paenibacillus. Each strain is considered to fit one genus, species, or strain if the similarity index value obtained meets the criteria. Based on Table 3, the five species of cellulolytic bacteria from the isolates belonged to the phylum Firmicutes: Bacillus wiedmannii, Bacillus marcorestinctum, Bacillus halotolerans, Paenibacillus zanthoxyli, and Bacillus hominis. The results of this study are in accordance with the findings of Wang et al.42 who isolated and screened five cellulose-degrading isolates from the gut of the grasshopper Yunnanacris yunnaneus.
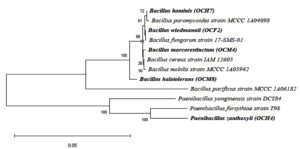
The phylogenetic tree of the cellulolytic bacterial isolates with 1,000 bootstrap repetitions is shown in Figure 4. The cellulolytic bacteria isolated from the gut of the grasshopper O. chinensis were closely related to B. wiedmannii FSL W8-0169, as indicated by an identity of 93.46%. Chantarasiri53 identified cellulolytic bacteria from freshwater wetlands and found B. wiedmannii strain W1401 in one of the isolates. Danial et al.54 reported that B. wiedmannii isolated from a cattle manure sample utilized sugar fruit peel waste efficiently to produce polyhydroxybutyrate.
Another isolate exhibited 94.03% identity to B. marcorestinctum strain LQQ. The latter was isolated from soil samples by Han et al.,55 who identified it as gram-positive, facultative anaerobic, rod-shaped bacterium. Effendi et al. 56 reported that B. marcorestinctum strain LQQ functioned as a biocontrol agent against spoilage in plants. Zhong et al.57 described the effect of B. marcorestinctum as a starter, which could improve the quality and safety of Yibin Yacai by changing the microbial community during fermentation.
A third isolate displayed 86.15% identity with B. halotolerans strain CR-119. Ouertani et al. 58 isolated B. halotolerans from a tannery wastewater and identified it as a keratinolytic bacterium. Yousef et al.59 recovered B. halotolerans from saline mud samples and demonstrated potential cellulase production. Wen et al.60 reported that B. halotolerans produced extracellular alkaline proteases, the activity of which was the highest after 12 h of incubation.
A fourth isolate exhibited 86.15% identity with P. zanthoxyli strain JH29. Ma et al.61 identified P. zanthoxyli as a novel nitrogen-fixing species isolated from the rhizosphere of Zanthoxylum simulans. The fifth isolate showed 95.79% identity with B. hominis strain BML-BC059. Several Bacillus species, such as B. cereus, B. licheniformis, B. megaterium, and B. subtilis, are known to be effective cellulolytic bacteria.33 The present study reports, for the first time, that B. marcorestinctum, P. zanthoxyli, and B. hominis possess cellulolytic activity in the grasshopper gut.
This study demonstrated the presence of lignocellulolytic activity of bacteria in the gut of O. chinensis. The 27 culturable bacterial isolates obtained exhibited cellulolytic and ligninolytic activity. This indicates that bacterial community in the gut of grasshopper plays in digesting food materials. Molecular identification based on the 16S rRNA gene sequence identified the five potential bacterial isolates as belonging to the genus Bacillus. The isolate with the highest cellulolytic ability was Bacillus wiedmannii (OCF2).
ACKNOWLEDGMENTS
The authors would like to thank the Universitas Sumatera Utara for funding the work under the scheme of Talenta Collaboration Research Grant of 2022.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by Universitas Sumatera Utara, contract number 356/UN5.2.3.1/PPM/KP-TALENTA/2022.
DATA AVAILABILITY
All data generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Pang J, Liu ZY, Hao M, Zhang YF, Qi QS. An isolated cellulolytic Escherichia coli from bovine rumen produces ethanol and hydrogen from corn straw. Biotechnol Biofuels. 2017;10:165-174.
Crossref - Dyartanti ER, Margono, Lestari IP, Putra MI, Pratiwi UI. Pretreatment ethanol from cellulosic. Equilibrium J Chem Eng. 2019;3(1):24-31.
Crossref - Dash S, Mubareka LH, Shil RK, Mojumder S, Sikdar D. Presence of cellulolytic and xylanolytic activities in the gut fluid of grasshopper Oxya velox. BioTechnologia (Pozn). 2019;100(3):199-207.
Crossref - Garg R, Srivastava R, Brahma V, Verma L, Karthikeyan S, Sahni G. Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome. Sci Rep. 2016;6(1):1-15.
Crossref - Islam F, Roy N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res Notes. 2018;11:445-450.
Crossref - Shanthi V, Roymon MG. Isolation, identification and partial optimization of novel xylanolytic bacterial isolates from Bhilai-Durg region, Chhattisgarh, India. Iranian J Biotechnol. 2018;16(3):200-212.
Crossref - Chaurasia B. Biological pretreatment of lignocellulosic biomass (water hyacinth) with different fungus for enzymatic hydrolysis and bio-ethanol production resource: advantages, future work and prospects. Acta Sci Agric. 2019;3(5):89-96.
- Wang L, Li C, Wang X, et al. Gut lignocellulose activity and microbiota in Asian longhorned beetle and their predicted contribution to larval nutrition. Front Microbiol. 2022;13:899865.
Crossref - Seo JK, Park TS, Kwon IH, Piao MY, Lee CH, Ha JK. Characterization of cellulolytic and xylanolytic enzymes of Bacillus licheniformis JK7 isolated from the rumen of a native Korean goat. Asian-Australas J Anim Sci. 2013;26(1):50-58.
Crossref - Setyoko H, Utami B. Isolation and characterization of cellulase enzymes cow’s liquid rumen for biomass hydrolysis. Proc Biol Educ Conf. 2016;13(1):863-867.
- Arfah RA, Natsir H, Atifah N, Zarkoni TR, Mahmud M. Isolation and characterization of soil termites (Macrotermes gilvus) cellulolytic bacteria and activity determination of cellulase enzyme on newsprint substrates. J Phys Conf Ser. 2019;1341(3):032037.
Crossref - Ejaz U, Sohail M, Ghanemi A. Cellulases: from bioactivity to a variety of industrial applications. Biomimetics. 2021;6(3):44-54.
Crossref - Raveendran S, Parameswaran B, Ummalyma SB, et al. Applications of microbial enzymes in food industry. Food Technol Biotechnol. 2018;56(1):16-30.
Crossref - Mason CJ, Jones AG, Felton GW. Co-option of microbial associates by insects and their impact on plant-folivore interactions. Plant Cell Environ. 2019;42(3):1078-1086.
Crossref - Gupta P, Samant K, Sahu A, Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol. 2012;(578925):1-5.
Crossref - Li J, Wang S, Zhao J, Dong Z, Shao T. Gut microbiota of Ostrinia nubilalis larvae degrade maize cellulose. Front Microbiol. 2022;13:816954.
Crossref - Sardar P, Sustr V, Chrooakova A, Lorenc F. Metatranscriptomic holobiont analysis of carbohydrate-active enzymes in the millipede Telodeinopus aoutii (Diplopoda, Spirostreptida). Front Ecol Evol. 2022;10:931986.
Crossref - Muratore M, Prather C, Sun Y. The gut bacterial communities across six grasshopper species from a coastal tallgrass prairie. PLOS One. 2020;15(1):1-14.
Crossref - Nelson K, Muge E, Wamalwa. Cellulolytic Bacillus Species Isolated from The gut of the desert locust Schistocerca gregaria. Sci African. 2020;11(1):e00665.
Crossref - Ferbiyanto A, Rusmana I, Raffiudin R. Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. Hayati. 2015;22(4):197-200.
Crossref - Sadhu S, Maiti TK. Cellulase production by bacteria: a review. Br Microbiol Res J. 2013;3:235-258.
Crossref - Simol CF, Chubo JK, King PJH, Ong KH, Chew C, Nawi K. Qualitative and molecular screening of potential ligninolytic microbes from termite (Coptotermes curvignathus) gut. Borneo J Resour Sci Technol. 2021;11(1):35-42.
Crossref - Sami A, Khalid M, Bilal S. Purification and characteristics of beta-1, 4-glucanase Occel9 of Oxya chinensis. Ann Res Rev Biol. 2014;4(24):4426-4444.
Crossref - Shil RK, Mojumder S, Sadida FF, Uddin M, Sikdar D. Isolation and identification of cellulolytic bacteria from the gut of three phytophagus insect species. Braz Arch Biol Technol. 2014;57(6):927-932.
Crossref - Upadhyaya SK, Manandhar A, Mainali H, et al. Isolation and characterization of cellulolytic bacteria from gut of termite. Rentech Symp Compendium. 2012;1(4):14-18.
- Fachrial E, Krisdianilo V, Harmileni, Nugroho INTT, Saryono. Isolation, characterization, activity test and molecular identification of thermophilic bacteria producing proteases from Dolok Tinggi Raja natural hot springs, North Sumatra, Indonesia. Biodiversitas. 2021;22(4):1725-1732.
Crossref - Bhakta CG. Isolation and screening of cellulolytic bacteria inhabiting different environment and optimization of cellulase production. Univers J Environ Res Technol. 2013;3(1):39-49.
- Lokhande S, Pethe AS. Isolation and screening of cellulolytic bacteria from soil and optimization of cellulase production. Int J Life Sci. 2017;5(2):277-282.
- Pham VHT, Kim J, Chang S, Chung W. Biodegradation of methylene blue using a novel lignin peroxidase enzyme producing bacteria, named Bacillus sp. React3, as a promising candidate for dye-contaminated wastewater treatment. Fermentation. 2022;8(5):1-15.
Crossref - Burlacu A, Cornea CP, Israel-Roming F. Screening of xylanase producing microorganisms. Res J Agric Sci. 2016;48(2):8-15.
- Irfan M, Safdar A, Syed Q, Nadeem M. Isolation and screening of cellulolytic bacteria from soil and optimization of cellulase production and activity. Turkish J Biochem. 2012;37(3):120-128.
Crossref - Miller GL. Use of dinitrosalisylic acid reagent for determination of reducing sugars. Anal Chem. 1959;31(3):426-428.
Crossref - Chantarasiri A. Novel halotolerant cellulolytic Bacillus methylotrophicus RYC01101 isolated from ruminant feces in Thailand and its application for bioethanol production. KMUTNB International Journal of Applied Science and Technology. 2014;7(3):63-68.
Crossref - Wusqy NK, Prayitno DI, Radjasa OK, Limantara L, Karwur FF. Investigation of bioactive compounds of sea anemone-associated bacteria and their molecular identification based on 16S rDNA. J Fish Sci. 2012;14(1):1-10.
- Dar MA, Shaikh AF, Pawar KD, et al. Evaluation of cellulose degrading bacteria isolated from the gut-system of cotton bollworm, Helicoverpa armigera and their potential values in biomass conversion. Peer J. 2021;9(11254):1-5.
Crossref - Ali HRK, Hemeda NF, Abdelaliem YF. Symbiotic cellulolytic bacteria from the gut of the subterranean termite Psammotermes hypostoma Desneux and their role in cellulose digestion. AMB Express. 2019;9:111.
Crossref - Boontanom P, Chantarasiri A. Diversity and cellulolytic activity of culturable bacteria isolated from the gut of higher termites (Odontotermes sp.) in Eastern Thailand. Biodiversitas. 2021;22(8):3349-3357.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Koubova A, Lorenc F, Horvathova T, Chrooakova A, Sustr V. Millipede gut-derived microbes as a potential source of cellulolytic enzymes. World J Microbiol Biotechnol. 2023;39(7):169-186.
Crossref - Willis JD, Klingeman WE, Oppert C, Oppert B, Jurat-Fuentes JL. Characterization of cellulolytic activity from digestive fluids of Dissosteira carolina (Orthoptera: Acrididae). Comp Biochem Physiol B Biochem Mol Biol. 2010;157(3):267-272.
Crossref - Abdullah R, Erfianti T, Pratama DA, Wijanarka. Isolation and screening of lactic acid bacteria from grasshopper gut as novel probiotic candidates to digest cellulose polymer. J Phys Conf Ser. 2019;1217(1):1-6.
Crossref - Wang JM, Bai J, Zheng FY, et al. Diversity of the gut microbiome in three grasshopper species using 16S rRNA and determination of cellulose digestibility. Peer J. 2020;8(10194):1-24.
Crossref - Dini IR, Wawan, Hapsoh, Sriwahyuni. Isolation and identification of cellulolytic and lignolytic bacteria from the gut Oryctes rhinoceros L. larvae decomposition of oil palm empty fruit bunches. Indones J Agric Res. 2018;1(2):193-203.
Crossref - Zhao L, Wang WQ, Xu SQ, Guan DL. The comparison of gut bacteria communities and the functions among the sympatric grasshopper species from the Loess Plateau. Front Microbiol. 2022;13:1-10.
Crossref - Saini A, Aggarwal NK, Yadav A. Isolation and screening of cellulose hydrolyzing bacteria from different ecological niches. Bioeng Biosci. 2017;5(1):7-13.
Crossref - Dineshkumar BA, Bhorkhataria BV, Jadhav PU, Palekar KS, Dhalkari M, Nalawade PM. Bacterial lignin peroxidase: a tool for biobleaching and biodegradation of industrial effluents. Univers J Environ Res Technol. 2012;2(1):58-64.
- Keerthana S, Kalaiselvi P, Maheshwari M, Kalaiselvi T. Isolation and screening of lignin and cellulose degrading proficient microbial strains from diverse biotic substrates based on qualitative traits. Int J Curr Microbiol Appl Sci. 2019;8(7):475-483.
Crossref - Gusmawartati, Agustian, Herviyanti, Jamsari. Isolation of cellulolytic bacteria from peat soils as decomposer of oil palm empty fruit bunch. J Trop Soils. 2017;22(1):47-53.
Crossref - Jimenez KR, Hernandez M. Isolation of fungi and bacteria associated with the guts of tropical wood-feeding coleoptera and determination of their lignocellulolytic activities. Int J Microbiol. 2015;(285018):1-11.
Crossref - Shi W, Ding SY, Yuan JS. Comparison of insect gut cellulase and xylanase activity across different insect species with distinct food sources. Bioenergy Res. 2011;4(1):1-10.
Crossref - Willis JD, Oppert B, Oppert C, Klingeman WE, Jurat-Fuentes JL. Identification, cloning, and expression of a GHF9 cellulase from Tribolium castaneum (Coleoptera: Tenebrionidae). J Insect Physiol. 2011;57(2):300-306.
Crossref - Sreena CP, Resna NK, Sebastian D. Isolation and characterization of cellulase producing bacteria from the gut of termites (Odontotermes and Heterotermes Species). Biotechnol J Int. 2015;9(1):1-10.
Crossref - Chantarasiri A. Diversity of cellulolytic bacteria isolated from a freshwater wetland reserve in Thailand and their cellulolytic activity. Appl Ecol Environ Res. 2020;18(4):5965-5983.
Crossref - Danial AW, Hamdy SM, Alrumman SA, Gad El-Rab SMF, Ahmed AMS, Hesham AEL. Bioplastic production by Bacillus wiedmannii AS-02 OK576278 using different agricultural wastes. Microorganisms. 2021;9(11):2395.
Crossref - Han Y, Chen F, Li N, Zhu B, Li X. Bacillus marcorestinctum sp. nov., a novel soil acylhomoserine lactone quorum-sensing signal quenching bacterium. Int J Mol Sci. 2010;11(2):507-520.
Crossref - Effendi Y, Khalisa A, Hadi AE, Pambudi A. Diversity, functional analysis and antagonistic potential of culturable-dependent soil bacteria from rhizosphere area of Fusarium oxysporum-infected banana tree. The 3rd International Conference on Life Sciences and Biotechnology (ICOLIB 2019).
- Zhong Y, Zou Y, Zheng Z, et al. Impact of inoculating with indigenous Bacillus marcorestinctum YC-1 on quality and microbial communities of yibin yacai (fermented mustard) during the fermentation process. Foods. 2022;11(22):3593.
Crossref - Ouertani R, Chouchane H, Mahjoubi M, et al. Feather degradation efficiency and hide dehairing ability of a new keratinolytic Bacillus halotolerans strain, isolated from a tannery wastewater. Appl Bionics Biomech. 2020;4(5):102-109.
Crossref - Yousef NMH, Aldaby ESE, Ali EE. Optimization of the cellulase enzyme production by Brevibacterium Halotolerans isolated from Wadi El-Natrun, Egypt. Assiut Univ J Bot Microbiol. 2020;49(1):14-33.
Crossref - Wen Y, Qiang J, Zhou G, Zhang X, Wang L, Shi Y. Characterization of redox and salinity-tolerant alkaline protease from Bacillus halotolerans strain DS5. Front Microbiol. 2022;13(935072):1-14.
Crossref - Ma Y, Zhang J, Chen S. Paenibacillus zanthoxyli sp. nov., a novel nitrogen-fixing species isolated from the rhizosphere of Zanthoxylum simulans. Int J Syst Evol Microbiol. 2007;57(4):873-877.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.