ISSN: 0973-7510
E-ISSN: 2581-690X
Klebsiella pneumoniae is a highly drug-resistant human pathogen responsible for a variety of serious infections. Integrons, mobile genetic elements capable of integrating antibiotic resistance genes, and the capsule are important virulence factors that increase bacteria resistance to phagocytosis and antimicrobial agents. Molecular typing is an effective tool for identifying the likely etiology of infection. This study aimed to investigate the presence of the rmpA, wcaG, intI1, intI2, and intI3 virulence genes in clinical Klebsiella pneumoniae isolates, and explore their molecular genotypes by using ERIC-PCR. Fifty Klebsiella pneumoniae strains were isolated from various specimens. Antimicrobial resistance was evaluated by using the disc diffusion method. Five genes were amplified by conventional PCR. Genotyping was performed molecularly by using ERIC-PCR. Forty-seven isolates were multi-drug resistant. In all, 18%, 36%, and 98% of the 50 K. pneumoniae isolates were positive for rmpA, wcaG, and intI1 genes, respectively; however, all isolates were negative for intI2 and intI3 genes. Dendogram analysis of the ERIC-PCR results showed 49 distinct patterns, arranged in five clusters. Our study demonstrates high levels of antibiotic resistance and virulence among clinical isolates of K. pneumoniae. Such resistance reflects a growing problem for public health. Further, the presence of integrons increases the horizontal spread of antibiotic resistance and virulence genes among bacterial isolates. The ERIC-PCR technique is an effective method for molecular typing and epidemiological studies of hospital-acquired infections.
Egypt, genotyping, integrons, Klebsiella pneumoniae, PCR
Klebsiella pneumoniae (K. pneumoniae) is a Gram-negative rod-shaped highly virulent bacterium, capable of causing serious diseases in humans including, pneumonia, liver abscesses, urinary tract infections, and life-threatening septicemia1-3. The bacterium is a major cause of nosocomial infections in Egypt4. K. pneumoniae currently manifests growing resistance worldwide to several important antimicrobial agents including, beta-lactam antibiotics, fluoroquinolones, and aminoglycosides. This increase in antimicrobial resistance constitutes an arising problem globally5. A primary mechanism for acquisition of resistance of K. pneumoniae is through integrons. These mobile genetic elements integrate and express antibiotic resistance genes. Integrons are carried and transferred by plasmids and transposons; facilitate spread by horizontal gene transfer within bacterial populations6. Five integron classes were defined based on difference in base sequences of integrase enzyme genes (int). Class 1 is the predominant among the five integron classes and is widely detected in Gram-negative bacteria, including K. pneumoniae. Structurally, class 1 integrons consist of two conserved regions: 3’ conserved segment and 5’ conserved segment, in addition to internal gene cassettes coding for antimicrobial resistance genes. Class 2 integrons can be detected occasionally in K. pneumoniae; in contrast, class 3 integrons are rarely documented. More than one hundred and thirty different cassettes that confer resistance against important antibiotic classes e.g.: ß- lactams, fluoroquinolones, aminoglycosides, and macrolides have been discovered7. The polysaccharide capsule is an important factor for bacterial virulence. The capsule describes bacterial mucoid phenotype and defines resistance to phagocytosis and host defense factors. For example, hypermucoid K. pneumoniae isolates are related to the incidence of invasive syndromes8. The plasmid gene rmpA activates transcription of the wzyKpK1 cps locus that in turn enhances capsule synthesis in K. pneumoniae2. Moreover, the wcaG virulence gene is gene located in transposable chromosomal region. This gene is responsible for synthesis of K. pneumoniae capsules, and is also helps in the conversion of mannose to fucose that may enhance the bacterial resistance to phagocytosis9.
Virulence analysis and molecular typing techniques are robust tools distinguish for identifying the probable source of infection, determining the genetic relationships in nosocomial infection outbreaks, and assisting in management and treatment of MDR K. pneumoniae infections10. Enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) is a molecular technique that is used to estimate genetic diversity among enterobacteriacae family members. ERIC sequences are 126 bp long, non-coding, and conserved. ERIC sequences occur in variable positions and numbers in bacteria, and the ERIC technique can be used to evaluate genetic differences among bacterial isolates. The use of the ERIC-PCR for investigation of the diversity of bacterial isolates is rapid, sensitive, and consistent11,12.
This study was aimed to investigate the presence of the rmpA, wcaG, and class 1, 2, and 3 integron virulence genes in clinical K. pneumoniae isolates, and to assess correlations among antibiotic resistance and virulence genes. Moreover, our isolates were molecularly genotyped by using the ERIC-PCR technique.
Sample collection, isolation, and identification
Seventy-five different samples were collected from patients at Damanhour General Hospital, El-Behira, Egypt from February to April 2018. The samples were collected from wound swabs, sputum, blood, endotracheal tube (ETT) aspirates, urine, and bedsore swabs. Samples were cultured on MacConkey agar plates for the detection of K. pneumoniae. Lactose fermenting mucoid colonies were Gram stained and subjected to several biochemical tests, including triple sugar iron agar, indole, methyl red, Voges Proskauer, citrate, oxidase, and catalase13. The identification of K. pneumoniae isolates was confirmed to the species level by using the automated vitek 2 system (Bio-Merieux, l’Etoile, France).
Antibiotic susceptibility testing
The antibiotic resistance was determined by using the standard disc agar diffusion technique according to Bauer et al.14 Fifteen commercially available antibiotic discs representing different antibiotic classes were used to assess resistance to ampicillin (AMP 10µg), amoxicillin/clavulanic acid (AMC 30µg), imipenem (IPM 10µg), cefuroxime (CXM 30µg), ceftazidime (CAZ 30µg), ceftriaxone (CRO 30µg), cefepime (FEP 30µg), tetracycline (TE 5µg), chloramphenicol (C 30µg), gentamycin (CN 10µg), aztreonam (ATM 30µg), amikacin (AKN 30µg) ciprofloxacin (CIP 5µg), cotrimoxthazole (SXT 25µg), and colistin (CT 10µg) (Oxoid® Ltd, England). Isolates were cultured on Mueller Hinton agar plates with antibiotic discs and incubated at 37°C for 24 hrs. The diameter of inhibition zones was measured in millimeters (mm). The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines15.
DNA extraction
DNA was extracted directly by using the boiling method with some modifications16. Briefly, 4_5 pure bacterial colonies of each isolate cultivated on nutrient agar plates were suspended in 500 μl of sterile water in a sterile Eppendorf tube, and heated at 95°C for 10 min. Bacterial suspensions were cooled on ice for 5 min, then centrifuged at 14,000 rpm for 5 min. The supernatants were transferred to sterile Eppendorf tubes and stored at_20°C.
Molecular detection of wcaG, rmpA, and class 1, 2, and 3 integrons genes by conventional PCR in our clinical K. pneumoniae isolates
All the DNA extracts were tested for five virulence genes (rmpA, wcaG, intI1, intI2, and intI3) by using a thermal cycler (Veriti, Applied Biosystems, Lincoln Foster City, California, USA) and five pairs of primers (Table 1). PCR amplicons were then resolved on 1.5 % agarose gel stained with ethidium bromide, and visualized via ultraviolet illumination. Cycling conditions were: initial denaturation at 94°C for 5 min; followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min, and a final extension at 72°C for 7 min9.
Table (1):
Primers used for the amplification of the five virulence genes9.
| The gene | The primers’ sequence | The amplicon size (bp) |
|---|---|---|
| rmpA | rmpA-F: ACTGGGCTACCTCTGCTTCA | 516 bp |
| rmpA-R: CTTGCATGAGCCATCTTTCA | ||
| wcaG | wcaG-F: GGTTGGGTCAGCAATCGTA | 169 bp |
| wcaG-R: ACTATTCCGCCAACTTT TGC | ||
| intI1 | intI1-F: CAGTGGACATAAGCCTGTTC | 160 bp |
| intI1-R: CCCGAGGCATAGACTG TA | ||
| intI2 | intI2-F: GTAGCAAACGAGTGACGAAATG | 789 bp |
| intI2-R: CACGGATATGCGA CAAAAAGGT | ||
| intI3 | intI3-F: GCCTCCGGCAGCGACTTTCAG | 979 bp |
| intI3-R: ACGG ATCTGCCAAACCTGACT |
Molecular genotyping of K. pneumoniae isolates by using ERIC-PCR
ERIC-PCR typing was performed by using the primer ERIC2 (5-AAGT AAGTGACTGGGGTGAGCG- 3) and with cycling conditions of: initial incubation at 94°C for 15 min., followed by 40 cycles at 94°C for 1 min, 37°C for 1 min, and final elongation at 72°C for 8 min. PCR products were resolved on 1.5% agarose gel stained with ethidium bromide, and visualized via ultraviolet illumination12. The gel electrophoresis results were analyzed by using the TotalLab Quant Analysis software (Version 1.0., TotalLab Ltd. United Kingdom) following the manufacturer’s instructions.
Statistical analysis
Correlations between the antibiotic resistance of the K. pneumoniae isolates, virulence genes, and ERIC genotypes were statistically determined by using Chi square test and Monte Carlo method by using the IPM SPSS software package version 20.0 (Armonk, NY:IBM Corp).
Fifty (66.7%) of 75 collected clinical samples were initially identified as Klebsiella species by their growth appearance on MacConkey agar plates and their morphological and biochemical characteristics. Mucoid lactose fermenting colonies appeared as Gram negative rods upon Gram staining. Biochemically, all isolates were oxidase negative, catalase positive, indole negative, methyl red negative, Voges Proskauer positive, and citrate positive. On triple sugar iron agar slants, all isolates produced acid butt and slant with gas and no H2S. These samples were confirmed as K. pneumoniae by using the automated vitek 2 system.
Twenty-seven (54%) of the fifty K. pneumoniae isolates were obtained from men and 23 (46%) from women. Five (10%) isolates were obtained from wound swabs, 12 (24%) from sputum, 9 (18%) from blood, 6 (12%) from endotracheal tube (ETT) aspirates, 16 (32%) from urine, and 2 (4%) from bedsore swabs.
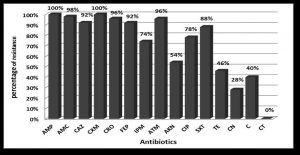
Forty-seven (94%) were multi-drug resistant (resistant to more than 3 different antibiotic classes). All (100%) isolates were resistant to ampicillin and ceforuxime. Isolates were highly resistant to amoxicillin/clavulanic acid, aztreonam, ceftriaxone, and ceftazidime. Conversely, all (100%) isolates were sensitive to colistin (Fig. 1). Eight resistance patterns were detected among the 50 K. pneumoniae isolates (Table 2).
Table (2):
Antibiotic resistance patterns of the 50 K. pneumoniae isolates.
The resistance pattern |
The antibiotics |
The number of isolates N (%) |
|---|---|---|
A1 |
CN-C-TE-IPM-FEP-ATM-AMP-AMC-CRO-CAZ-CXM-AKN-CIP-SXT |
7 (14%) |
A2 |
IPM-CN-FEP-ATM- AMP-AMC-CRO-CAZ-CXM-AKN-CIP-SXT |
5 (10%) |
A3 |
C-TE-IPM-FEP-ATM-AMP-AMC-CRO-CAZ-CXM- CIP-SXT |
4 (8%) |
A4 |
IPM-FEP-ATM-AMP-AMC-CRO-CAZ-CXM-AKN-CIP-SXT |
11(22%) |
A5 |
TE-IPM-FEP-ATM-AMP-AMC-CRO-CAZ-CXM -CIP-SXT |
4 (8%) |
A6 |
C-TE-FEP-ATM-AMP-AMC-CRO-CAZ-CXM-SXT |
2 (4%) |
A7 |
FEP-ATM-AMP-AMC-CRO-CAZ-CXM-SXT |
2 (4%) |
A8 |
AMP-AMC-CXM |
2 (4%) |
Further, nine (18%) of the 50 K. pneumoniae isolates were positive for the rmpA gene and 18 (36%) for the wcaG. Forty-nine (98%) isolates were positive for the intI1 gene (Fig. 2 and 3); however, all (100%) K. pneumoniae isolates were negative for intI2 and intI3 genes. Only one isolate (no.47) was negative for all genes. The remaining forty-nine isolates showed four virulence patterns (Table 3).
Table (3):
Virulence patterns of the 49 K. pneumoniae isolates.
The virulence pattern |
Isolate codes |
Number of isolates N (%) |
Genes |
|---|---|---|---|
V1 |
4, 9 and 10 |
3 (6%) |
positive for rmpA, wcaG and intI1 genes |
V2 |
26, 33, 38, 41, 42 and 46 |
6 (12%) |
positive for rmpA and intI1genes |
V3 |
1, 3, 5, 6, 7, 8, 12, 16, 17, 18, 19, 21, 22, 24 and 31 |
15 (30%) |
positive for wcaG and intI1genes |
V4 |
2, 11, 13, 14, 15, 20, 23, 25, 27, 28, 29, 30, 32, 34, 35, 36, 37, 39, 40, 43, 44, 45, 48, 49 and 50 |
25 (50%) |
positive for intI1gene |
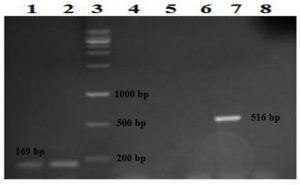
Fig. 2. Detection of the rmpA and wcaG genes on 1.5% agarose gel following amplification with PCR. Lanes 1 and 2: two isolates positive for the wcaG gene detected at 169 bp, lane 3: the DNA ladder, lanes 4, 5, 6, and 8: negative isolates and lane 7: one isolate positive for the rmpA gene detected at 516 bp.
Significant correlations were found between the intI1 gene and resistance to aztreonam and ceftriaxone (p-value ≤ 0.05), and between the rmpA gene and resistance to gentamicin and amikacin (p-value ≤ 0.05). No significant correlations were observed between the wcaG gene and resistance to any antibiotic (p-value < 0.05).
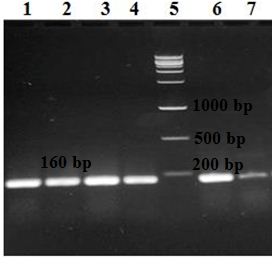
Fig. 3. Detection of the intI gene on 1.5% agarose gel following amplification with PCR. Lanes 1, 2, 3, 4, 6, and 7: six isolates positive for the intI gene detected at 160 bp and lane 5: the DNA ladder.
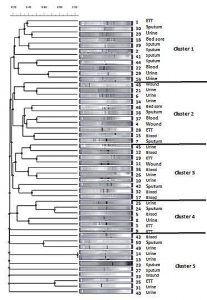
The 49 genotypically positive isolates were genotyped by using ERIC-PCR, followed by analysis with TotalLab Quant Analysis software (Version 1.0., TotalLab Ltd. United Kingdom). This analysis used the Dice method for comparison and the UPGMA method for clustering. Dendogram analysis of K. pneumoniae isolates showed forty-nine distinct patterns, arranged in five clusters. These clusters contained 11, 11, 10, 6, and 11 of the 49 K. pneumoniae isolates, respectively (Fig. 4).
Fig. 4. Dendrogram generated with Dice coefficient and the UPGMA clustering method, showing the genetic similarity among K. pneumoniae isolates by Enterobacterial Repetitive Intergenic Consensus (ERIC) genotyping.
A significant correlation was found between the presence of the wcaG gene and ERIC genotypes (p-value ≤ 0.05), however, no such correlation was found for rmpA or intI1 (p-value < 0.05).
Klebsiella pneumoniae is a dangerous pathogen that causes serious wound, urinary tract, respiratory tract infections, meningitis, and bacteremia. K. pneumoniae is a common cause of nosocomial infections, and serious outbreaks are reported in hospitals along with increased morbidity and mortality rates. The uncontrolled use of antibiotics for treating K. pneumoniae infections has resulted in increased bacterial resistance and the emergence of MDR strains12,17.
Forty-seven (94%) of our 50 K. pneumoniae isolates were MDR. In contrast, lower percentages (54%, 38%, 81.5%, and 90%) of MDR were reported in other studies17-19. The high percentage of MDR in our isolates may be due to overuse of antibiotics in Egypt and the incidence of class 1 integrons in 98% of our isolates. These integrons spread antibiotic-resistant genes by horizontal gene transfer. High resistance percentages ranging from 92% to 100% were exhibited for penicillins and cephalosporins, including ampicillin, ceforuxime, amoxicillin/clavulanic acid, ceftazidime, ceftriaxone, and cefepime, and lower percentages were reported in other studies17,19,20. High level of resistance was also displayed in this study to imipenem, co-trimoxazole, aztreonam, and ciprofloxacin; however, lower percentages were reported by other studies17,19,20. Moderate incidence of resistance (54%, 46%, and 40%) was exhibited for amikacin, tetracycline, and chloramphenicol, respectively, in this study. Such incidence is similar to previous reports19,20. Alternatively, 28% of our isolates were resistant to gentamycin; however, a higher percentage was reported by Farivar et al.19. Finally, all isolates in this study were sensitive to colistin; in contrast, Farivar et al.19 reported a lower percentage (83%). Overall, the incidence of antibiotic resistance among our isolates was high compared to previous studies.
We found that 18 (36%) out of isolates were positive for the wcaG gene by conventional PCR. Lower percentages (6.5% and 23%) were reported by Zhang et al. in China and by Derakhshan et al. in Iran9,21. Nevertheless, Zaki et al., in Egypt (22) reported a higher percentage (50%).
Similarly, we found that 9 (18%) of isolates were positive for the rmpA gene. A similar percentage (17%) was reported by Guo et al. in China (23). In contrast, lower percentages (1.6%, 7%, and 10%) were reported by Zhang et al., Derakhshan et al., and Zaki et al., however, a higher percentage (46%) was reported by Tan et al. in Singapore9,21,22,24.
Finally, forty-nine (98%) out of our 50 K. pneumoniae isolates were positive for the intI1 gene. Lower percentages (74%, 69%, and 53%) were reported by Derakhshan et al., Zaki et al., and Liao et al.; in contrast, all (100%) isolates of Firoozeh et al. in Iran were positive for this gene7,9,22,25.
None of isolates was positive for intI2, consistent with the result reported by Zaki et al. in Egypt22. Nonetheless, Derakhshan et al. in Iran and Liao et al. in China reported that 1% and 2% of their isolates, respectively, were positive for class 2 integrons9,25. In addition, Firoozeh et al. in Iran reported that 37% of their isolates were positive for this gene7. Similarly, none of our isolates was positive for intI3; this finding matches the results reported by Derakhshan et al., Zaki et al. and Firoozeh et al.7,9,22.
Significant correlations were found between the intI1 gene and resistance to aztreonam and ceftriaxone (p-value ≤ 0.05). In contrast, Derakhshan et al. reported a positive association between the intI1 gene and resistance to cefotaxime, ceftriaxone, ceftazidime, amoxicillin-clavulanic acid, aztreonam, ciprofloxacin, tobramycin, tetracycline, co-trimoxazole, gentamicin, and cefepime. In our study, significant correlations were also found between presence of the rmpA gene and resistance to gentamicin and amikacin (p-value ≤ 0.05); however, Derakhshan et al. reported a positive association between this gene and resistance to amoxicillin-clavulanic acid, tobramycin, and gentamicin. In contrast, we observed no significant correlations between the wcaG gene and resistance to any antibiotic (p value < 0.05); nevertheless, Derakhshan et al. reported a positive association between the gene and resistance to all antibiotics assessed, except imipenem9. Significant correlations confirm that virulence genes rmpA and intI1 in the bacterial genome often co-occur antimicrobial resistance.
The diversity in the ERIC patterns observed in the study may reflect the non-clonal distribution of virulent K. pneumoniae strains. Our data support the results of Wasfi et al. as their Dendrogram analysis of ERIC genotyping revealed 21 distinct patterns among 28 isolates divided into three clusters (A–C) containing 12/28, 9/28, and 7/28 of the isolates, respectively12. Similarly, Zhang et al. used ERIC-PCR to reveal 60 different distinct patterns among their 62 strains, and Mehr et al. reported 32 different ERIC profiles among their 35 isolates21,26. Such genetic variation among K. pneumoniae strains was also reported by other studies in Iran, Taiwan, Russia, and Algeria27-30.
In conclusion, our study demonstrates a high incidence of antibiotic resistance and virulence among Klebsiella pneumoniae clinical isolates, reflecting the continuing threat to public health that needs to be followed up continuously. The presence of class 1 integrons increases the risk of spreading antibiotic resistance genes and virulence factors through horizontal gene transfer. Moreover, the ERIC-PCR technique is a very effective method for molecular typing and epidemiological studies of nosocomial infections.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Prof. Dr. S designed the experiments. Dr. K. Dr. M and Dr. H performed the experiments. Dr. K and Dr. M analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
FUNDING
None.
ETHICS STATEMENT
The current research has followed the accepted principles of ethical conduct by the Research Ethics Committee of the Faculty of Pharmacy, Damanhour University, and it has been approved. Informed consent from the parents of young patients was obtained prior to undertaking testing and molecular investigation of their specimens.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Abbott SL. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and Other Enterobacteriaceae. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (Eds.), Manual of Clinical Microbiology. 9th ed. 698-711. Washington, USA. 2007.
- Vila A, Cassata A, Pagella H, et al. Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol J. 2011;5:107-113.
Crossref - Minarini LAR, Gales AC, Palazzo IC, Darini ALC. Prevalence of community-occurring extended spectrum β-lactamase-producing Enterobacteriaceae in Brazil. Curr Microbiol. 2007;54(5):335-341.
Crossref - Daef EA, Elsherbiny NM. Clinical and microbiological profile of nosocomial infections in adult intensive care units at Assiut University hospitals, Egypt. J Am Sci. 2012;8(12):1239-1250.
- Ferreira RL, da Silva BCM, Rezende GS, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-Lactamase encoding genes in a brazilian intensive care unit. Front Microbiol. 2019;9:3198.
Crossref - Carattoli A. Importance of integrons in the diffusion of resistance. Vet Res. 2001;32(3-4):243-259.
Crossref - Firoozeh F, Mahluji Z, Khorshidi A, Zibae M. Molecular characterization of class 1, 2 and 3 integrons in clinical multi-drug resistant Klebsiella pneumoniae isolates. Antimicrob Resist Infect Control. 2019;8:59.
Crossref - Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. rmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192(12):3144-3158.
Crossref - Derakhshan S, Peerayeh SN, Bakhshi B. Association between presence of virulence genes and antibiotic resistance in clinical Klebsiella Pneumoniae isolates. Lab Med. 2016;47(4):306-311.
Crossref - Sachse S, Bresan S, Erhard M, et al. Comparison of multilocus sequence typing, RAPD, and MALDI-TOF mass spectrometry for typing of β-lactam-resistant Klebsiella pneumoniae strains. Diagn Microbiol Infect Dis. 2014;80(4):267-271.
Crossref - Barus T, Hanjaya I, Sadeli J, Lay BW, Suwanto A, Yulandi A. Genetic diversity of Klebsiella spp. isolated from tempe based on enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR). Hayati J Biosciences. 2013;20(4):171-176.
Crossref - Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929.
Crossref - Finegold SM, Baron EJ. Bailey and Scott’s Diagnostic Microbiology. The C.V. Mosby company: St. Louis. 1986.
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-396.
Crossref - CLSI. Performance standards for antimicrobial susceptibility testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and laboratory standards institute. 2019.
- Queipo-Ortun MI, Colmenero JDD, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15(2):293-296.
Crossref - Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13(Suppl 11):20.
Crossref - Cepas V, Lopez Y, Munoz E, et al. Relationship between biofilm formation and antimicrobial resistance in Gram-negative Bacteria. Microb Drug Resist. 2019;25(1):72-79.
Crossref - Farivar SA, Nowroozi J, Eslami G, Sabokbar A. RAPD PCR profile, antibiotic resistance, prevalence of armA gene, and detection of KPC enzyme in Klebsiella pneumoniae isolates. Can J Infect Dis Med. 2018;2018:6183162.
Crossref - Manjula NG, Math CG, Nagshetty K, Patil SA, Gaddad SM, Shivannavar CT. Antibiotic susceptibility pattern of ESbetaL producing Klebsiella pneumoniae isolated from urine samples of pregnant women in Karnataka. JCDR. 2014;8(10):8-11.
- Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and Genotypic characterization of Klebsiella pneumoniae isolated from retail Foods in China. Front Microbiol. 2018;9:289.
Crossref - Bakr AO, Zaki A. Molecular study of Klebsiella Pneumoniae virulence genes from patients with hospital acquired sepsis. Clin Lab. 2019;65(1).
Crossref - Guo S, Xu J, Wei Y, Xu J, Li Y, Xue R. Clinical and molecular characteristics of Klebsiella pneumoniae ventilator-associated pneumonia in mainland China. BMC Infect Dis. 2016;16(1):608.
Crossref - Tan TY, Ong M, Cheng Y, Ng LSY. Hypermucoviscosity, rmpA, and aerobactin are associated with community-acquired Klebsiella pneumoniae bacteremic isolates causing liver abscess in Singapore. J Microbiol Immunol. 2019;52(1):30-34.
Crossref - Liao W, Lia D, Liu F, et al. Distribution of integrons and phlogenetic groups among highly virulent serotypes of Klebsiella pneumoniae in a chinese tertiary hospital. J Glob Antimicrob Re. 2020;21:278-284.
Crossref - Mehr PV, Shokoohizadeh L, Mirzaee M, Savari M. Molecular typing of Klebsiella pneumoniae isolates by enterobacterial repetitive intergenic consensus (ERIC)-PCR. Infection Epidemiology and Microbiology (IEM). 2017;3(4):112-116.
- Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in russian hospitals. Antimicrob. Agents Chemother. 2003;47(12):3724-32.
Crossref - Lagha N, Abdelouahid DE, Hassaine H, Robin FE, Bonnet R. First characterization of CTXM- 15 and DHA-1-lactamases among clinical isolates of Klebsiella pneumoniae in Laghouat Hospital, Algeria. Afr J Microbiol Res. 20148(11):1221-1227.
- Seifi K, Kazemian H, Heidari H, et al. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur J Microbiol. 2016;9(1):e30682.
Crossref - Yan J-J, Hsueh P-R, Lu J-J, et al. Extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes among clinical isolates of Escherichia coli and Klebsiella pneumoniae from seven medical centers in Taiwan. Antimicrob Agents Chemother. 2006;50(5):1861-1864.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.