ISSN: 0973-7510
E-ISSN: 2581-690X
The current study was performed to identify the genotypes related to Klebsiella pneumonia isolated from urinary tract infection (UTI) of humans and pneumonia (PMN) of sheep in Al-Qadisiyah province, Iraq. 140 samples of (80 UTI and 60 post-mortems identified PMN) were collected and processed using traditional and molecular techniques. Following isolation of the microorganism, pure colonies were introduced into steps of a polymerase chain reaction (PCR) confirming method that targeted the 16S rRNA and the virulence-related capsular (magA and k2A) genes and partial gene sequencing (PGS) which focused on the 16S rRNA gene only. Primary results of the traditional techniques (TTs) highly revealed the presence of K. pneumonia in 11 (13.75%) and 18 (30%) of the UTI and the PMN samples, respectively. However, rest samples showed the presence of different bacteria with no bacterial growth in others. This was confirmed by PCR results that strongly uncovered the identity of the K. pneumoniae in the positive samples. Using specific primers for Maga and k2A genes, PCR results revealed full percentage based presence of the k2A gene in the UTI and the PMN samples with less, 3/7 (43%) and 4 (100%) in the UTI and the PMN samples, respectively, presence of the magic gene. Moreover, PGS findings confirmed the accuracy of the TT and the PCR results plus recognized specific local strain identities that differently aligned with some of the worlds isolates on a phylogenetic tree. Current findings provide valuable information about virulence status of the K. pneumoniae isolates. in Al-Qadisiyah province, Iraq.
Genotyping, Klebsiella pneumoniae, PCR, sequencing
Klebsiella pneumoniae is a bacterial microorganism that is considered as an opportunistic pathogen inducing influential human and animal related diseases1. Although K.pnneumoniae bacteria are present normally in/on the human and animal mucosal membranes of different body systems such as the urinary system or the respiratory system2, the microorganism causes different infections in humans generated through various initiators such as nosocomial, direct or indirect contact with infected animals, and infected dairy products3, and some of these infections are infected wounds, UTIs, bronchus and lung infections, bacteremia, meninges-related infections,differently-body-located abscesses3,4. K.pnneumoniae bacteria were isolated from mastitis in cows5 and upper respiratory tract infection (URTI) and pyogenic lung infections in sheep6. This wide range of bacterial presence and pathogenicity introduces important health problems accompanied by significantly increasing multi-antibacterial resistance (MAR) dilemmas7. One of the pathogenicity strength points of the bacterium is provided via the presence of the virulence factor; capsule, enhancing major self-protection against environmental and serum factor and the phagocytic killing activities generating variable degrees of infection severities according to the types of the capsular (K) antigens discovered that exceeded 80 components8–11. For pyogenic related infections in Asian countries, K1 andK2 are recognized to be the major capsular antigenic types present12. The well-known strains with low severity of K. pneumoniae have been evolved showing more virulent strains with increased mucoviscosity properties causing extremely severe infections such as abscesses of liver in immuno competent and immuno compromised young people inducing Gram positive pyogenic bacteria-like bodily dissemination with increasing worldwide infection rates that may alarm for a huge crisis especially with their elevating capabilities of acquiring new MAR properties11,13–15. The current exploring study was performed to identify the genotypes, using the capsular virulence factors of magA and k2A genes, related to Klebsiella pneumoniae isolated from UTIs of humans and PMN of sheep in Al-Qadisiyah province, Iraq.
Sampling
The work was initiated by collecting of 140 samples (80 UTI from patients attended Al-Diwaniyah Teaching Hospital and 60 post-mortem identified PMN from sheep in a slaughterhouse, Al-Qadisiyah Provonce, Iraq) samples. These samples were processed using traditional and molecular based techniques. MaCconkey’s agar was used to cultivate all the samples overnight incubation at 37°C that was followed by overnight eosin methylene blue (EMB) agar and chrom agar based sub-cultivating of all the lactose fermenting colonies at 37°C. Using morphological and biochemical tests accompanied by the API20E System, a primary diagnosis of the K. pneumonia was performed following standards from16.
After isolation of the microorganism had done, pure colonies were introduced into steps of a polymerase chain reaction (PCR) confirming method that targeted the 16S rRNA and the virulence-related capsular (magA and k2A) genes and partial gene sequencing (PGS) which focused on the 16S rRNA gene only.
Extraction of DNA and PCR method
Extraction of the DNA was enhanced recruitingBosphore® Bacterial DNA Extraction spin kit (Bosphore®, Germany) with depending on the protocol accompanied the kit. The obtained DNA was tested for its quality and quantity using a NanoDrop.
The PCR amplification method was launchedusing 20µl as a total volume for the PCR reaction. The primers shown in table (1) were used for this purpose. A DNA template at 5µl was added to a mixture of 1.5µl (10 pmol/ µl)of each of the forward and the reverse primers, 250µM of dNTP, 30mM of KCl, 1.5mM of MgCl2, and Taq DNA polymerase at 1 unit. The total volume was then reached by adding water made for molecular biology use. The conditions for the athermocycler (TECHNE, USA) used are shown in table (1). Then, the products of the PCR at 10µl/each were electrophoresed through a 1.5%-agarose gel pre-treated with 0.5µl/ml ethidium bromide. After the process was done, the gel containing the amplicons was screened and photographed using a UV imager and a specific camera, respectively. For the sequencing, 11 positive samples were sent out to do PGS.
Table (1):
The sequences of the primers and the conditions of the thermocycler followed in this study
| Gene | Sequence5’→3’ | Denaturation (°C-min) | Annealing(°C-s) | Extension(°C-min) | Final extension (°C-min) | Cycle No. | Ampilcon size (bp) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Initial | Main | |||||||||
| 16S rRNA | F | AGAGTTTGATCCTGGCTCAG | 95-4 | 94-1 | 60-40 | 72-1 | 72-10 | 30 | 1500 | (17) |
| R | GGTTACCTTGTTACGACTT | |||||||||
| magA | F |
GGTGCTCTTTACATCATTGC
|
95-5 | 94-1 | 51-60 | 72-1 | 72-10 | 35 | 1282 | (18) |
| R | GCAATGGCCATTTGCGTTAG | |||||||||
| K2A | F | CAACCATGGTGGTCGATTAG | 95-5 | 94-1 | 58-60 | 72-1 | 72-10 | 30 | 523 | (19) |
| R | TGGTAGCCATATCCCTTTGG | |||||||||
Traditional techniques
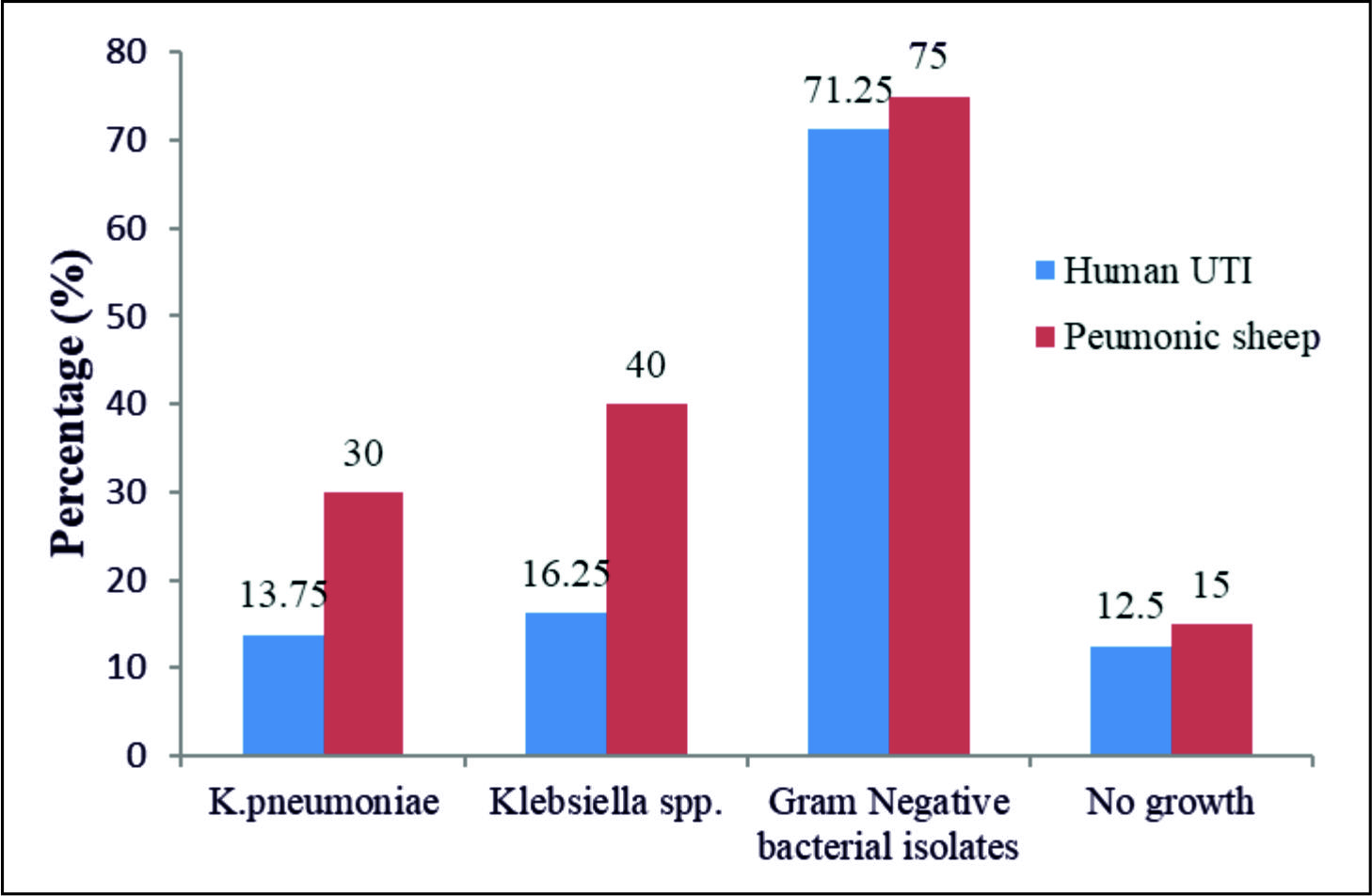
The primary results of the traditional techniques (TTs) highly revealed the presence of K.pneumoniae in 11/80(13.75%) and 18/60(30%) of the UTI and the PMN samples, respectively. However, the rest of the samples showed the presence different bacteria with no bacterial growth in 25 (17.9%), 10 UTI and 15 PMN, samples, table 2 and Fig. 1.
Table (2):
Numbers of bacterial isolates obtained from UTI and PMN samples
| Source of samples | N | Bacterial isolates (No. (%)) | No bacterial growth (No. (%)) | X2(Pvalue) | ||
|---|---|---|---|---|---|---|
| K.pneumoniae | Klebsiella spp. | Gram –ve | ||||
| Human | 80 | 11(13.75) | 13(16.25) | 57(71.25) | 10(12.5) | 96.35(0)S |
| Sheep | 60 | 18(30) | 24(40) | 45(75) | 15(25) | 37.44(0)S |
| Total | 140 | 29(20.71) | 37( 26.42) | 102(72.85) | 25(17.85) | 124.18(0)S |
| X2 (pvalue) | 3.379(0.066)NS | 9.94(0.002)S | 0.244(0.621)NS | 3.65(0.056)NS | ||
S: Significant alterations (p<0.05); NS: Non-significant changes(p˃0.05).
Result of PCR
The results of the PCR method strongly uncovered the identity of the K.pneumoniae in the positive samples according to the amplified piece of the 16S rRNA gene at size 1500bp Fig. 2.
Using the specific primers mentioned above for the magAand k2A genes, the PCR results revealed full percentage based presence of the k2Agene, 523bp, in the UTI and the PMN samples with less, 3/7 (43%) and 4 (100%) in the UTI and the PMN samples, respectively, presence of the magA gene,1282bp,table 3 and Fig. 3, 4, and 5.
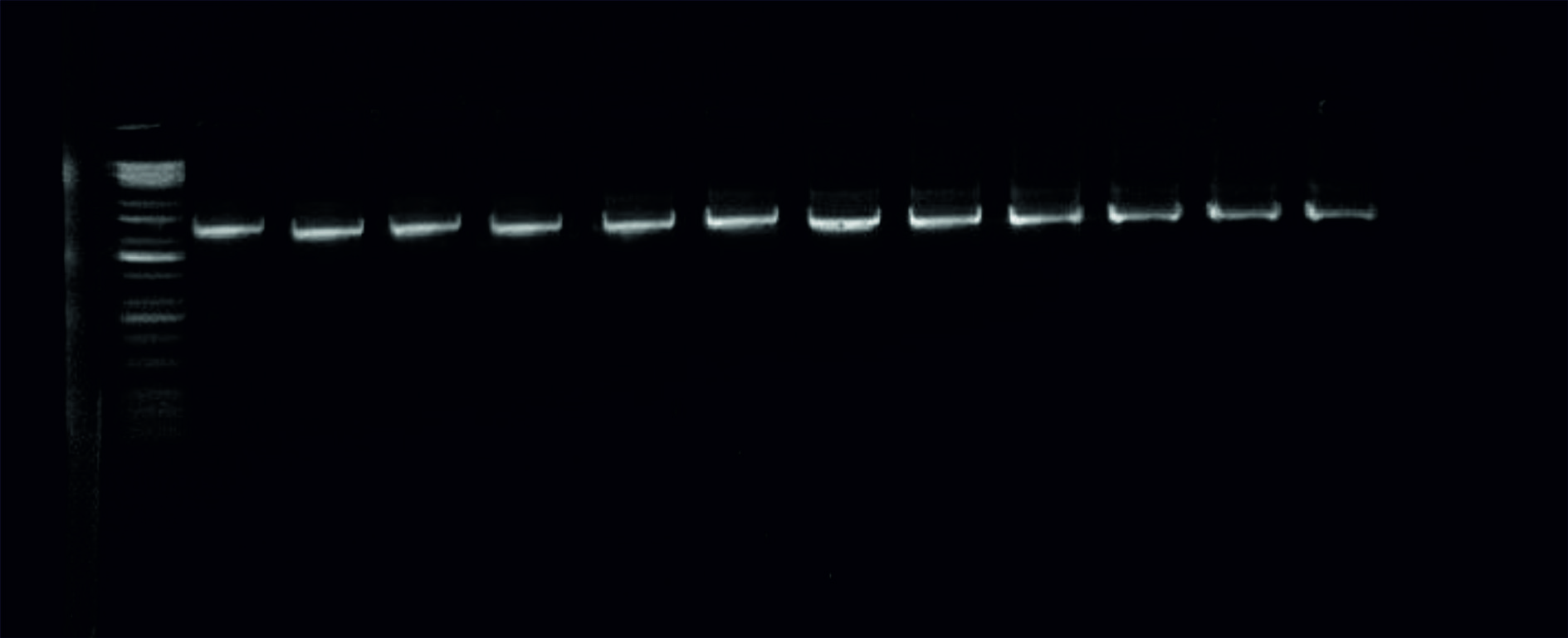 Fig. 2. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting the 16S rRNA gene at 1500bp. The size of the ladder is 10Kbp.
Fig. 2. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting the 16S rRNA gene at 1500bp. The size of the ladder is 10Kbp. 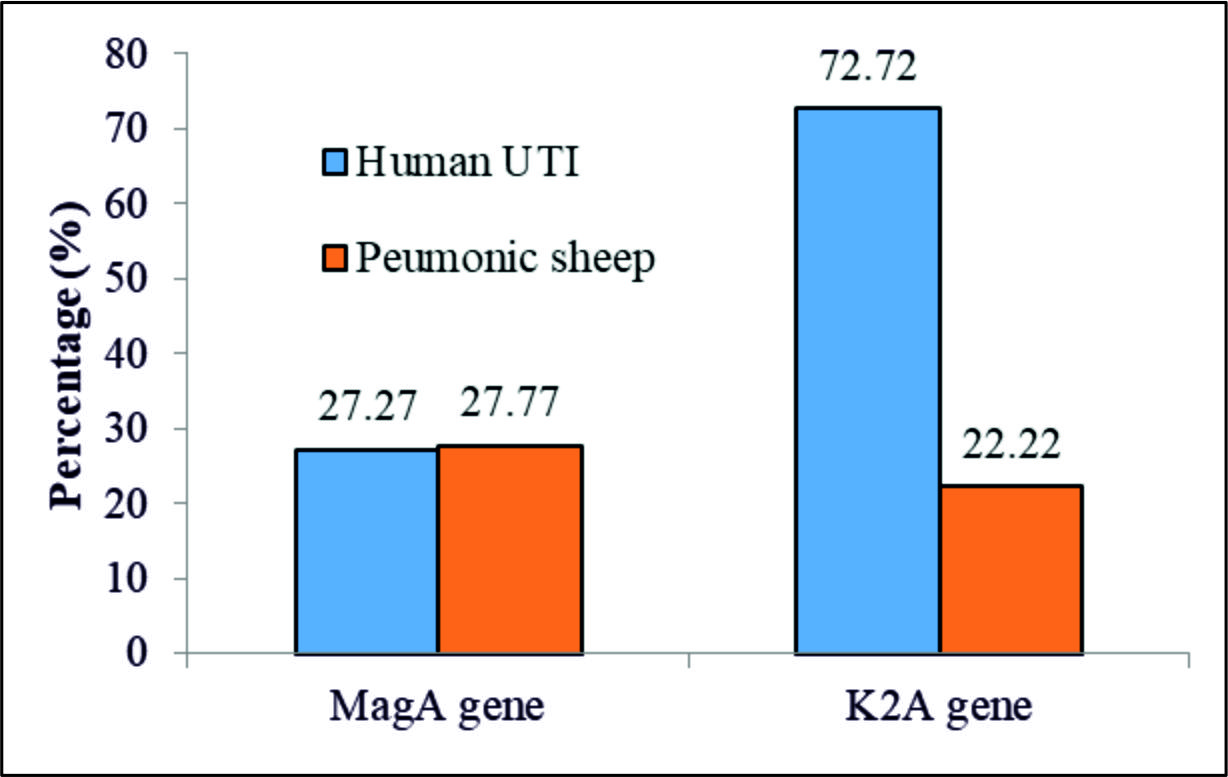 Fig. 3. PCR results regarding the magA and the k2A genes in the positive K. pneumonia samplesfrom UTI and PMN.
Fig. 3. PCR results regarding the magA and the k2A genes in the positive K. pneumonia samplesfrom UTI and PMN.  Fig. 4. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting thek2A gene, 523bp. The size of the ladder is 10Kbp
Fig. 4. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting thek2A gene, 523bp. The size of the ladder is 10Kbp 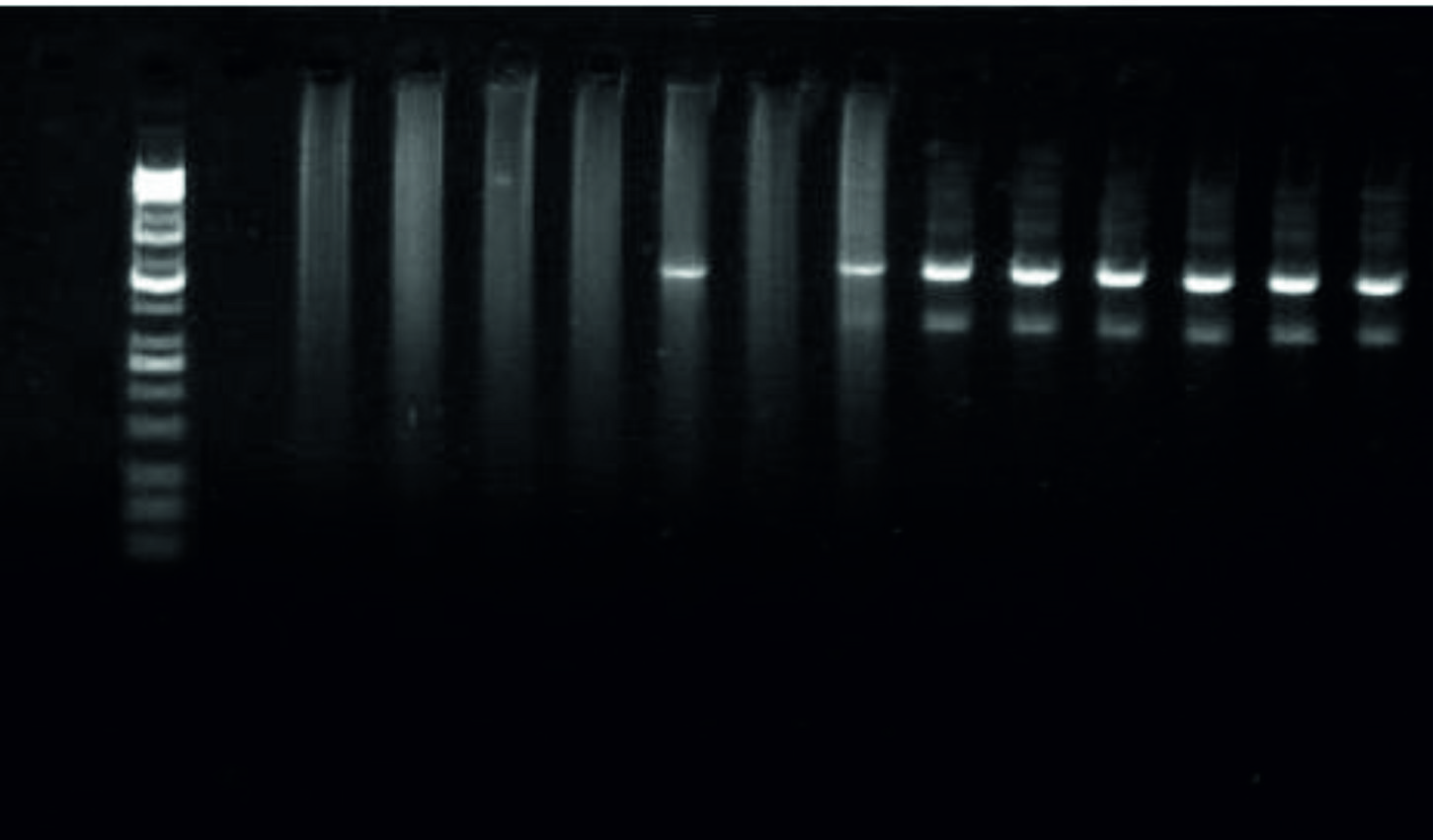 Fig. 5. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting themagA gene, 1282bp. The size of the ladder is 10Kbp
Fig. 5. Image of the electrophoresed agarose gel. The PCR products of K.pneumoniaetargeting themagA gene, 1282bp. The size of the ladder is 10Kbp Table (3):
Numbers of positive bacterial isolates to K. pneumoniae magAand k2A genes obtained from UTI and PMN samples
| Gene name | No. of isolates and percentage (%) | X2(p value) | |||
|---|---|---|---|---|---|
| UTI of humans (N=11) | Pneumonic sheep(N=18) | ||||
| Positive | % | Positive | % | ||
| MagA(K1serotype) | 3 | 27.27 | 5 | 27.77 | 0.001(0.976)NS |
| k2A(k2 serotype) | 8 | 72.72 | 4 | 22.22 | 7.18(0.007)S |
| X2 (P value) | 4.545(0.033)S | 0.148(0.700)NS | |||
S: Significant alterations (p<0.05); NS: Non-significant changes(p˃0.05).
Klebsiella pnneumoniaeis a bacterial microorganism that is considered as an opportunistic pathogen inducing influential human and animal related diseases1. There is a big increase in the emerging of hyper virulent strains with increased mucoviscosity properties causing extremely severe infections such as abscesses of liver in immunocompetent and immunocompromised young people with elevating capabilities of acquiring new MAR properties that increase the complexity of the problems facing these strains11,13–15.
In the current work, the findings revealed more frequent presence (100%) of the k2Agene in the UTI and the PMN samples than that for the case of the magA gene. These results agree with20 who identified that the k2A genotype was more dominant than that for the case of the magA genotype in samples of human urine, exudate, bronchial wash, endo-tracheal aspirates, fluids of the pleura, and blood.However, these findings showed dissimilarities with results obtained from human samples in different countries such as K1 appeared to take over the occurrence rate (68.9%) results in east China with less appearance rate (20%) of the K2 based serotype from abscesses in the liver21 with showing the same results in a study from Germany22. In a study performed in Taiwan, the serotype K1 was revealed in a higher occurrence rate, 63.4%,that that for the case of K2, 14.2%, of K. pneumonia isolates from liver abscess23. In Japan, it has been detected that the rate for the K1 serotype occurrence was lower than that regarding the K2 serotype of K. pneumonia isolates from liver abscesses24 which shows similar to the current study results. These serotypes were also identified from the globe different regions such as from Europe, Africa, Australia,North America, and the Middle East25. The results from the respiratory tracheal swabs showed matching to the findings observed by26 who identified the presence of lung invasion signs after performing the PM investigations such as lung hemorrhage and congestions detected in dead minks and mice with higher occurrence rates of the K2 serotype than that regarding the K1 serotype.
For the case of the phylogenetic study, the results recognized specific local strain identities that were differently aligned with some of the world isolates on the phylogenetic tree, Fig. 6.
 Fig. 6. The phylogenetic tree based on the 16SrRNA gene partial sequencing. The tree was built up using the Maximum Composite Likelihood method that was applied to MEGA X software.
Fig. 6. The phylogenetic tree based on the 16SrRNA gene partial sequencing. The tree was built up using the Maximum Composite Likelihood method that was applied to MEGA X software. For those strains aligned together, these results might indicate similar ancestors as they may have evolved and transmitted from similar regions of the world; however, some strains have aligned in distinguished branches of the tree suggesting a complete evolving process resulting in new line of strains that are not similar to other strains from the selected world regions27. The current findings provide valuable information about the current virulence status of the K.pneumoniae isolates in Al-Qadisiyah province, Iraq.
Acknowledgements
None.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
All authors have made substantial contribution to the work and approved it for publication.
Funding
None.
Data Availability
All data generated during the study are included in the manuscript.
Ethics Statement
The study was undertaken after obtaining approval from the Institutional Research and Ethics Committee.
- Mansour AMA, Zaki HM, Hassan NA, Al-Humiany AA, Mansour AMA, Zaki HM, et al. Molecular characterization and immuno-protective activity of capsular polysaccharide of klebsiella pneumoniae isolated from farm animals at taif governorate. Am. J. Infect. Dis., 2014.
Crossref - Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, Thi Tran HK, et al. Klebsiella pneumoniae Oropharyngeal Carriage in Rural and Urban Vietnam and the Effect of Alcohol Consumption. Kluytmans J. 2014; editor.
Crossref - Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol. Biol. Rev., 2016. https://doi.org/10.1128/MMBR.00078-15
- Fang C-T, Lai S-Y, Yi W-C, Hsueh P-R, Liu K-L, Chang S-C. Klebsiella pneumoniae Genotype K1: An Emerging Pathogen That Causes Septic Ocular or Central Nervous System Complications from Pyogenic Liver Abscess. Clin. Infect. Dis., 2007; 45(3): 284-93.
Crossref - Munoz MA, Bennett GJ, Ahlstrצm C, Griffiths HM, Schukken YH, Zadoks RN. Cleanliness Scores as Indicator of Klebsiella Exposure in Dairy Cows. J Dairy Sci., 2008.
Crossref - Azizi S, Korani FS, Oryan A. Pneumonia in slaughtered sheep in south-western Iran: pathological characteristics and aerobic bacterial aetiology. Vet Ital, 2013.
- Bassetti M, Righi E, Carnelutti A, Graziano E, Russo A. Multidrug-resistant Klebsiella pneumoniae/ : challenges for treatment, prevention and infection control. Expert Rev. Anti. Infect. Ther., 2018; [Internet]. Oct 3 [cited 2019 May 3]; 16(10): 749-61.
Crossref - Podschun R, Pietsch S, Hצller C, Ullmann U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol., 2001; [Internet]. Jul 1 [cited 2019 May 3];67(7):3325-7.
Crossref - Broberg CA, Palacios M, Miller VL. Klebsiella: a long way to go towards understanding this enigmatic jet-setter, 2014.
Crossref - Pan Y-J, Lin T-L, Chen Y-H, Hsu C-R, Hsieh P-F, Wu M-C, et al. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. Forestier C, editor, PLoS One, 2013;
Crossref - Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, Cha C-J, et al. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front Cell Infect Microbiol., 2017; [Internet]. Nov 21 [cited 2019 May 3]; 7.
Crossref - Juan C-H, Chuang C, Chen C-H, Li L, Lin Y-T. Clinical characteristics, antimicrobial resistance and capsular types of community-acquired, healthcare-associated, and nosocomial Klebsiella pneumoniae bacteremia. Antimicrob Resist Infect Control, 2019.
Crossref - Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis., 2016.
Crossref - Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence, 2013.
Crossref - Kotekani, L; Kotigadde, S/ ; Nair B. T. Capsular Serotyping and Antimicrobial Drug Resistance of Klebsiella pneumoniae Strains Isolated from a Tertiary Care Hospital, Southern India. J. Diagnost Rese., 2018; 12(8): 1-5.
Crossref - MacFaddin. Biochemical tests for identification of medical bacteria. 3rd ed. Baltimor, USA: Williams and Wilkins, 2000; 64-67 p.
- Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic Analysis of Aquaspirillum magnetotacticum Using Polymerase Chain Reaction-Amplified 16S rRNA-Specific DNA. Int. J. Syst. Bacteriol., 1991.
Crossref - Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp Med., 2004.
Crossref - Chuang Y, Fang C, Lai S, Chang S, Wang J. Genetic Determinants of Capsular Serotype K1 of Klebsiella pneumoniae Causing Primary Pyogenic Liver Abscess. J. Infect. Dis., 2006.
Crossref - Remya P, Shanthi M, Sekar U. Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae. J. Lab. Physicians, 2018.
Crossref - Qu T, Zhou J, Jiang Y, Shi K, Li B, Shen P, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis., 2015.
Crossref - Bilal S, Volz MS, Fiedler T, Podschun R, Schneider T. Klebsiella pneumoniae -Induced Liver Abscesses, Germany. Emerg. Infect. Dis., 2014.
Crossref - Fung C-P, Chang F-Y, Lee S-C, Hu B-S, Kuo BI-T, Liu C-Y, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis, 2002.
Crossref - Seo R, Kudo D, Gu Y, Yano H, Aoyagi T, Omura T, et al. Invasive liver abscess syndrome caused by Klebsiella pneumoniae with definite K2 serotyping in Japan: a case report. Surg Case Reports, 2016.
Crossref - Jian-li W, Yuan-yuan S, Shou-yu G, Fei-fei D, Jia-yu Y, Xue-hua W, et al. Serotype and virulence genes of Klebsiella pneumoniae isolated from mink and its pathogenesis in mice and mink, 2017.
Crossref - Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol., 2018.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.