ISSN: 0973-7510
E-ISSN: 2581-690X
Silver nanoparticles possess unique physical, chemical, and biological properties which find myriad applications but the random application of silver nanoparticles may arise many problems particularly on benefits biotechnological agent T. harzianum which serves as bio-control and growth promoting agents for many crop plants. The results indicated that AgNPs possess antifungal properties against T. harzianum and the antifungal activity increased with increasing their concentrations. Electrophoretic analysis was performed to determine the effect of AgNPs on T. harzianum protein profile. Protein migration in this study demonstrated that nanoparticles caused changes in the protein profiles. Two bands completely disappeared after treatment with 100 and 200 ppm of AgNPs with molecular weights 140, 27 and 21.0 kDa. The genotoxicity exhibited by AgNPs was demonstrated by DNA fragmentation post treatment particularly at high concentrations of the nanoparticles. The fragmented bands increased with increasing AgNPs concentrations. 2, 5 and 7 fragmented bands with different molecular bands were detected at 50,100 and 200 ppm of AgNPs respectively. Three bands with molecular weight 1000,900 and 750 bp were detected at 100 and 200 ppm of AgNPs. Also, the result showed that AgNPs treated fungal cells both accumulated more intracellular glucose and trehalose than the compound-untreated cells. Also, the result showed that T. harzianum treated with AgNPs increased extracellular glucose and trehalose than the compound-untreated cells as a result of cell wall damage.
Genotoxicity, molecular response, Trichoderma harzianum, silver nanoparticles
Nanomaterials are defined by their small size (< 100 nm) and their novel physical, chemical and biological properties, which are progressively applied on numerous research and economical fields. However, the rapid progress in nanoscience has not been accompanied by enough information regarding its toxicity. In the past ten years, particularly the past three years, a large number of scientific papers have been published in an attempt to understand various aspects of the hazards of silver nanoparticles (AgNPs). Several reviews have also dealt with the exposure, environmental fate, and in vivo and in vitro toxicities of AgNPs (Lynch and Dawson 2008; Reidy et al., 2013; Yu et al., 2013). The unique properties of AgNPs make them ideal for numerous technologies, including biomedical, optical materials, optical, and antimicrobial applications (Kim et al., 2007; Choi et al., 2008).
Trichoderma harzianum used in several application, it has the ability to minimize the severity of fungal and bacterial plant diseases by inhibiting plant pathogens through their high antagonistic and mycoparasitic potential (Rosa et al., 2012; Hussein et al., 2017). Moreover, T. harzianum can solubilize several plant nutrients such phosphorus compounds (Janardan et al., 2011), Certain Trichoderma spp. have beneficial impacts on plant growth and decrease sensitive to both biotic and a biotic stresses (Elshahawy et al., 2017).
The antimicrobial effects of silver have been known for numerous years. Over the past several years, the use of silver or silver salts as key components to control microbial proliferation has become increasingly popular (Quadros and Marr 2011). As a broad-spectrum antimicrobial agent, AgNPs are widely used in medical and consumer products, including household antiseptic sprays and antimicrobial coatings for medical devices (Faunce and Watal 2010; Zhi-Kuan et al., 2016). Hwang et al. (2012) demonstrated for the first time that AgNPs promotes apoptosis in Candida albicans through phosphatidylserine exposure. AgNPs may also cause DNA damage and damage replication ability (Morones et al., 2005). Shunmugadevi and Palanisamy (2016) cleave DNA of Escherichia coli completely compared to untreated DNA with AgNPs. As a result, it can be said that the ability of these compounds for the DNA cleavage may be considered as a major reason for the inhibitory effect of them on the growth of the pathogenic organisms. According to Gopinath et al., (2010) AgNPs can interact with membrane proteins and activate signaling pathways, leading to repress cell proliferation. Cytotoxic and/or genotoxic effect of AgNPs and the ability to induce various chromosomal aberrations in mammalian cells, bacteria and root meristematic cells of different plants were reported in numerous studies (Patra et al., 2007; Kumari et al., 2009; Sahar et al., 2014). In addition, treatment with an AgNPs suspension may decrease the replication ability of bacterial DNA and inactivate the cellular proteins (Feng et al., 2000).
Moreover, nanoparticles as well as AgNPs are also known to induce oxidative stress in microbes which will eventually lead to the killing of microbes. It has been previously reported that increased reactive oxygen species (ROS) production due to AgNPs damage membranes, forming free radicals with a powerful microbiocidal action (Wu et al., 2014). Study on AgNPs interactions with the phosphate groups and nitrogenous bases in DNA was earlier reported (Sheikpranbabu et al., 2010). Some investigations showed that OH ion is active structures and the strongest antimicrobial elements; these free radicals attack to interior DNA of fungus and bacterium, and cause apoptosis in cells (Naghsh et al., 2012). Another mechanism of antifungal activity of AgNPs was reported (Keuk-Jun 2009; Siddhanta et al., 2016), where the analysis of glucose and trehalose release, during AgNPs exposure, suggests that it may be one of several intracellular components released during membrane disruption by AgNPs. This study aimed to explore the genotoxic and DNA-damaging potential of AgNPs using bioapplicable agent T. harzianum to avoid AgNPs using in pest control in the presence of this fungus.
Biotechnological agent and culture conditions
Biotechnological agent Trichoderma harzianum was used a commercially available product (Plant guard produced by El-Nasser Com. Egypt ). The rate of mycelial growth was measured in colonies grown in Petri dishes containing PDA supplemented with different concentration (50,100 and 200 ppm) of AgNPs (<100nm ) at 30°C for different incubation periods. The average diameter of each colony was measured daily to record the growth and growth inhibition %.
Protein gel-electrophoresis
Five grams of each dried fungus treated with AgNPs or untreated were ground in a volume of 0.1 ml sample buffer (sodium dodecyl sulfate) cracking solution. Extracts were added in 1.5 cm eppendorf centrifuge tube according to Laemmli (1970). Homogenates were heated at 95°C for 5 min then briefly centrifuged at 12,000 rpm to pellet cellular debris. The resulting supernatants (total protein extracts) were stored at -70°C until analysis by Polyacrylamide Gel Electrophoresis. The extract was separated by electrophoresis on 1mm thick 12.5% acrylamide slab gels. Gels were stained with Coomassie blue at Regional Center for Mycology and Biotechnology (RCMB), Al-Azhar University, Egypt.
DNA fragmentation of T. harzianum
T. harzianum cultivated at different concentrations of AgNPs at 30°C for 6 days. At the end of the incubation, the harvested, mycelia were treated with 200 µl cell lysis buffer (50 mM TrisHCl, pH 8.0, 10 Mm ethylene diamine tetra acetic acid, 0.1 M sodium chloride, and 0.5% sodium dodecyl sulphate) for 1 h at 37°C. The lysate was incubated with 0.2 mg/ml proteinase K at 40°C for 2 h. After completion of incubation, the sample was centrifuged for 10 min at 10,000 rpm. The aqueous portion, containing the DNA was transferred to new Eppendorf tube. DNA fragmentation assay was performed on 1% agarose gel (Vahdati and Sadeghi, 2013). Genomic DNA was loaded into the slots of 1% agarose gel containing 1 µg/ml Ethidium bromide, at a constant power supply of 80 volts for one hour. The gel was visualized under UV transilluminator and documented.
Determining released glucose and trehalose
T. harzianum after cultivation in Potato Dextrose Broth Medium was harvested, washed three times with PBS, and then 5 ml of the T. harzianum sores suspension (2 x 104 spores), containing different concentrations 50, 100 and 200 ppm of AgNPs, were incubated for 5 hrs at 30°C in Phosphate buffered saline (PBS). The control was incubated without AgNPs. The spores were settled by centrifugation (at 12,000 rpm for 15 min). The supernatants were transferred to a new tube. Released glucose and trehalose-containing supernatants were added to 0.05 units of trehalase. After 1 hr of enzymatic reaction at 37°C, the reaction suspension was mixed with water and 16% DNS reagent (3, 5-dinitrosalicylic acid 1%, NaOH 2%, sodium potassium tartrate 20%) was added. For the reaction of glucose with the DNS reagent, the mixture was boiled for 5 min and cooled. Color formations were measured at 525 nm. The Intracellular trehalose was measured according to a modified previously described method (Pedreno et al., 2007).
The inhibitory effect of AgNPs to living organisms has been investigated in a number of studies. But many studies failed to describe the behavior of nanoparticles in the particular biological media. Therefore the purpose of our study was to investigate the toxicity AgNPs to commercial biocontrol agent T. harzianum. AgNPs treatment with 50,100 and 200 ppm concentration resulted in about 29.48, 66.66 and 73.33% inhibition percentage of T. harzianum respectively at 6 days of incubation period (Table 1 and Figure 1). Although the growth increased at 8 days but the inhibition percentage of growth increased this may due to resistance of T. harzianum to AgNPs. Therefore, the results suggested that maximum inhibition was obtained at treated with 200 ppm concentration of AgNPs at all tested incubation periods. Similarly, a recent publication showed that AgNPs had antifungal effects against fungi (Abdlghany 2013, Sahar 2014; Zhi-Kuan et al., 2016, Abdlghany et al., 2017).
Table (1):
Growth and inhibition % of T. harzianum exposure to AgNPs at different incubation periods
| Incubation period (Day) | AgNPs concentration ppm | |||||||
|---|---|---|---|---|---|---|---|---|
| Control | 50 | 100 | 200 | |||||
| Growth | Inhibion % | Growth | Inhibion % | Growth | Inhibion % | Growth | Inhibion % | |
| 2 | 1.2±0.2 | 0 | 0.8±0.8 | 33.33 | 0.0±0.0 | 100 | 0.0±0.0 | 100 |
| 4 | 4.5±0.5 | 0 | 3.2±0.4 | 28.88 | 2.2±0.4 | 51.11 | 1.4±0.4 | 68.88 |
| 6 | 7.8±0.2 | 0 | 5.5±0.5 | 29.48 | 2.5±0.5 | 66.66 | 2.0±0.1 | 73.33 |
| 8 | 8.2±0.4 | 0 | 7.0±0.2 | 14.63 | 2.8±0.2 | 65.85 | 2.4±0.7 | 70.73 |
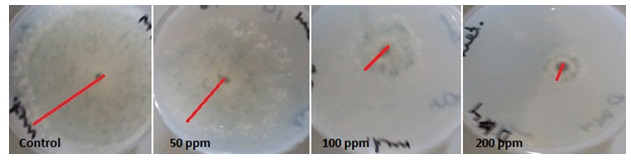
Fig. 1. Growth of T. harzianum exposure to different concentrations of AgNPs at 6 days incu-bation period
SDS-PAGE gel electrophoresis was carried out to monitor the change in gene expression of T. harzianum exposed to different concentration to AgNPs (Figure 2). The average total number of bands per lane induced in T. harzianum was 18 with molecular weight ranging from 150 to 12.5 kDa (Table 2). There were numerous protein bands present were detected in treated and in untreated T. harzianum by AgNPs. Only two bands completely disappeared after treatment with 100 and 200 ppm of AgNPs with molecular weights 140, 27 and 21.0 kDa (Table 2). The amount of expression of proteins in each treatment changed compared with the control, where the band intensity of each treatment was changed as compared with their control (Figure 2).These results agreed with a previous study observed that nanometer-sized silvers possess different properties, which might come from structural and physiological changes (Nel et al., 2006; Sahar 2014). It is evident that AgNPs directly interacts with macromolecular structures of living cells and exerts an active influence on their metabolism. They can also cause damage to the nuclear DNA by altering the chemical structure of the nucleotide bases and the deoxyribosyl backbone (Cooke et al., 2003). The cytotoxic effects of silver are the result of active physicochemical interaction of silver atoms with the functional groups of intracellular proteins, as well as with the nitrogen bases and phosphate groups in DNA.
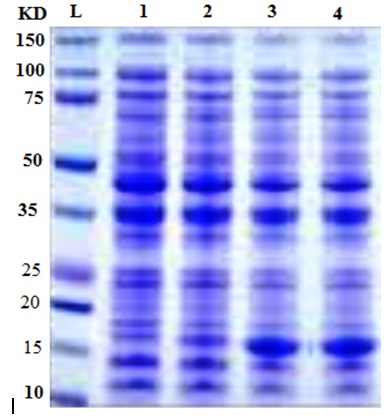
Fig. 2. Protein profile of T. harzianum at different concentrations of silver nanoparticles.
Table (2):
Protein bands (kd) detected at different concentrations of silver nanoparticles
| Band No. | Molecular weight of protein bands (kd) at different concentrations of | |||
|---|---|---|---|---|
| Control | 50 ppm | 100 ppm | 200 ppm | |
| 1 | 12.5 | 12.5 | 12.5 | 12.5 |
| 2 | 14.8 | 14.8 | 14.8 | 14.8 |
| 3 | 16.7 | 16.7 | 16.7 | 16.7 |
| 4 | 18.5 | 18.5 | 18.5 | 18.5 |
| 5 | 19.3 | 19.3 | 19.3 | 19.3 |
| 6 | 21.0 | 21.0 | – | – |
| 7 | 25.0 | 25.0 | 25.0 | 25.0 |
| 8 | 24.0 | 24.0 | 24.0 | 24.0 |
| 9 | 27.0 | 27.0 | 27.0 | 27.0 |
| 10 | 35.0 | 35.0 | 35.0 | 35.0 |
| 11 | 45.0 | 45.0 | 45.0 | 45.0 |
| 12 | 50.0 | 50.0 | 50.0 | 50.0 |
| 13 | 60.0 | 60.0 | 60.0 | 60.0 |
| 14 | 72.5 | 72.5 | 72.5 | 72.5 |
| 15 | 75.0 | 75.0 | 75.0 | 75.0 |
| 16 | 100 | 100 | 100 | 100 |
| 17 | 140 | 140 | – | – |
| 18 | 150 | 150 | 150 | 150 |
Agarose electrophoresis of genomic DNA from fungus exposed to silver nanoparticles revealed fragmented DNA compared to control (Figure 3). Agarose gel electrophoresis showed intact DNA bands with the untreated DNA (control), where no significant damage occurred. In contrast, fungs treated with different concentrations of silver nanoparticles showed a dose-dependent induction of DNA strand break, characterized by increased DNA fragmentation (Figure 3). In the present study, the genotoxicity exhibited by AgNPs was demonstrated by DNA fragmentation post treatment particularly at high concentrations of the AgNPs. The fragmented bands increased with increasing AgNPs concentrations. 2, 5 and 7 fragmented bands with different molecular bands were detected at 50,100 and 200 ppm of AgNPs respectively. Three bands with molecular weight 1000,900 and 750 bp were detected at 100 and 200 ppm of AgNPs (Table 3). Such genotoxic activities of nanoparticles were reported earlier (Supriyo et al., 2014) where degree of DNA degradation was directly proportional to the concentration of AgNPs. DNA smear of the NPs treated fungus showed that the AgNPs treated cells exhibited extensive double strand breaks of various sizes, thereby yielding a smear like appearance (lane 2,3 and 4), while the DNA of control cells exhibited no breakage (lane 1). DNA fragmentation assay revealed that the AgNPs showed genotoxic effect in a dose dependent manner. These results suggest that the biocidal effect of AgNPs may occur by direct chemical damage to DNA. These results are in accordance with the findings by Sobhy et al., (2012), showing action of AgNPs on DNA of Alternaria solani. Recently, Mukesh (2016) reported that NPs caused damage of E. coli genomic DNA in a dose dependent manner. Sobhy et al. (2012), stated that he mechanism of AgNPs antifungal activity may be related to damaging the membrane lipid bilayer, leading to intracellular ion efflux resulting in cell death. Also, accumulation of AgNPs in the cell nuclei and interaction with DNA may lead to cell death.
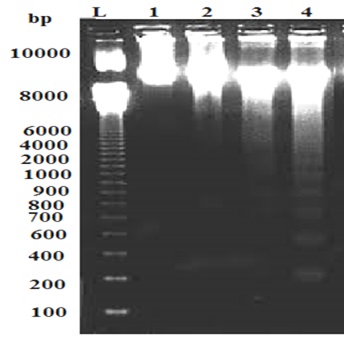
Fig. 3. DNA Fragmentation Assay of T. harzianum at different concentrations of AgNPs. M: Ladder; 1: Control; 2:50 ppm; 3: 100 ppm; 4:200 ppm AgNPs.
Table (3):
DNA Fragmented Bands of T. harzianum at different concentrations of AgNPs
| Treatment with AgNPs | Molecular Weight of Fragmented Bands | |
|---|---|---|
| Ladder | 10 K bp | |
| Control | 9600 bp | |
| 50 ppm | Band 1 | 9100 bp |
| Band 2 | 8200 bp | |
| 100 ppm | Band 1 | 8900 bp |
| Band 2 | 7800 bp | |
| Band 3 | 1000 bp | |
| Band 4 | 900 bp | |
| Band 5 | 750 bp | |
| 200 ppm | Band 1 | 8800 bp |
| Band 2 | 7900 bp | |
| Band 3 | 1000 bp | |
| Band 4 | 900 bp | |
| Band 5 | 750 bp | |
| Band 6 | 500 bp | |
| Band 7 | 300 bp | |
In the current study, measuring the glucose and trehalose released was used to assess the ability of AgNPs to disturb the integrity of the plasma membrane of fungal cells. Kim et al., (2009) reported that AgNPs exhibited potent antifungal effects, probably through destruction of membrane integrity. The result showed that AgNPs treated fungal cells both accumulated more intracellular glucose and trehalose than the compound-untreated cells (Figure 4). The rate of accumulation increased at highest concentration of AgNPs and reached to maximum (26.50 µg/mg) at 100ppm of AgNPs. At the same time, fungal cells also increased extracellular glucose and trehalose than the compound-untreated cells. Release of glucose and trehalose may explained as a result of cell wall damage, such changes were reported in previous studies (Elbein et al., 2003; Kim et al., 2009; Niazi et al., 2011). Recently Siddhanta et al., (2016) found that perturbation of membrane by AgNPs leads to the generation of glucose and trehalose which indicate that they are the intracellular components of the membrane.
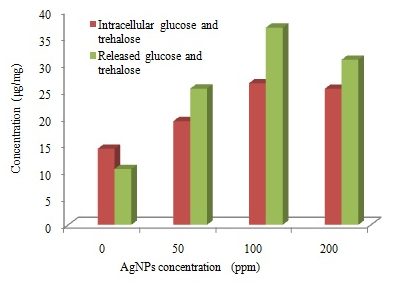
Fig. 4. Trehalose and glucose concentrations (μg/mg) detected in T. harzianum exposed to different concentrations of AgNPs
In the present study AgNPs have exhibited antifungal and genotoxic effect on T. harzianum and therefore this paper suggest that AgNPs, because of the harmful effects on environment, should be applied less in the presence this biotechnological agent T. harzianum.
- Abdelghany TM, Aisha MH Al-Rajhi, Mohamed A Al Abboud, Alawlaqi MM, Ganash A Magdah; Eman AM Helmy, Ahmed S Mabrouk. Recent Advances in Green Synthesis of Silver Nanoparticles And Their Applications: About Future Directions. A Review BioNanoSci., 2017 7, DOI 10.1007/s12668-017-0413-3
- Abdelghany TM. Stachybotrys chartarum: a novel biological agent for the extracellular synthesis of silver nanoparticles and their antimicrobial activity. Indonesian J of Biotechnology, 2013; 18 (2): 75-82.
- Choi O , Deng KK, Kim N Jr, Ross L, Rao YS, Hu Z. The inhibitory effects of silver nanoparticles, silver ions and silver chloride colloids on microbial growth. Water Res. 2008; 42(12):3066-3074.
- Cooke MC, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage:mechanisms, mutation, and disease. FASEB J, 2003; 17(10):1195-214.
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I. & Carroll, D. New insights on trehalose: a multifunctional molecule. Glycobiology, 2003; 13(4) 17-27.
- Elshahawy IE., Nehal S., Abd-El-Kareem F. & Morsy A. Biocontrol of onion white rot by application of Trichoderma species formulated on wheat bran powder. Archives of Phytopathology and Plant Protection, 2017; 50:(3-4) 150-166.
- Faunce T. and Watal A. Nanosilver and global public health: international regulatory issues Nanomedicine, 2010; 5: 617–632.
- Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000; 52(4):662–668.
- Gopinath P, Gogoi SK, Sanpuic P. Signaling gene cascade in silver nanoparticle induced apoptosis. Colloids Surf B, 2010; 77: 240-245.
- Hussein A. S., Basheer N. J. , Ali D. K. , Iman S. S. , Abdalsalam AA. Effect of Biocontrol, Physical Control and Compost on Tomato Plants that Infected with Fusarium wilt under Greenhouse Conditions. World Journal of Agricultural Research, 2017; 5(1): 5-8.
- Hwang I., Juneyoung L. , Hwang JH, Keuk-Jun K. and Dong G. L. Silver nanoparticles induce apoptotic cell death in Candida albicans through the increase of hydroxyl radicals. FEBS Journal, 2012; 279(7): 1327–1338.
- Janardan Yadav, Jay Prakash Verma and Kavindra Nath Tiwari, Plant Growth Promoting Activities of Fungi and their Effect on Chickpea Plant Growth. Asian Journal of Biological Sciences, 2011; 4: 291-299.
- Keuk-Jun K.; Woo S. S. ; Bo K. S. ; Seok-Ki M.; Jong-Soo C.; Jong G. K.; Dong G. L. Antifungal activity and mode of action of silver nano-particles on Candida albicans, Biometals, 2009; 22: 235-242.
- Kim YK, Lee YS, Jeong DH, Cho MH. Antimicrobial effect of silver nanoparticles. Nanomedicine, 2007; 3(1):95–101.
- Kim, K.J.; Sung, W.S.; Suh, B.K.; Moon, S.K.; Choi, J.S.; Kim, J.G. & Lee, D.G. Antifungal activity and mode of action of silver nano-particles on Candida albicans. Biometals, 2009; 22(2): 235-242.
- Kumari M, Mukherjee A, Chandrasekaran N. Genotoxicity of silver nanoparticles in Allium cepa. Sci Total Environ., 2009; 407(19):5243–5246.
- Laemmli UK, Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature (London), 1970; 227:680-685.
- Lynch I, Dawson KA. Protein nanoparticle interactions (review). Nanotoday, 2008; 3:40- 7.
- Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramý´rez JT, The bactericidal effect of silver nanoparticles. Nanotechnology, 2005; 6: 2346-2353.
- Mukesh S. Elucidation of biogenic silver nanoparticles susceptibility towards Escherichia coli: an investigation on the antimicrobial mechanism . IET Nanobiotechnol., 2016; 10(5): 276-280.
- Naghsh N , Safari M.and Hajmehrabi P. Comparison of nanosilver inhibitory effects growth between Aspergillus niger and E. coli. Indian Journal of Science and Technology, 2012; 5(S3): 2448-2450.
- Nel, A.E., T. Xia, L. Madler and N. Li. Toxic potential of materials at the nanolevel. Science, 2006; 311: 622-627.
- Niazi, J. H., Sang, B. I., Kim, Y. S., & Gu, M. B. Global Gene Response in Saccharomyces cerevisiae exposed to silver nanoparticles. Applied Biochemistry and Biotechnology, 2011; 164(8): 1278–1291.
- Patra HK, Banerjee S, Chaudhuri U, Lahiri P, Dasgupta AK. Cell-selective response to gold nanoparticles. Nanomedicine, 2007; 3(2):111–119.
- Pedreno Y, González-Párraga P, Martínez-Esparza M, Sentandreu R, Valentin E, Argüelles JC. Disruption of the Candida albicans ATC1 gene encoding a cell-linked acid trehalase decreases hypha formation and infectivity without affecting resistance to oxidative stress. Microbiology, 2007; 153:1372-1381.
- Quadros M.E. and Marr L.C. Silver nanoparticles and total aerosols emitted by nanotechnology-related consumer spray products. Environ Sci Technol, 2011; 45: 10713-10719.
- Reidy B, Haase A, Luch A, Mechanisms of silver nanoparticle release, transformation and toxicity: a critical review of current knowledge and recommendations for future studies and applications. Materials, 2013; 6:2295-350.
- Rosa H., Ada V., llan C. and Enrique M. Plant-beneficial effects of Trichoderma and of its genes. Microbiology, 2012; 158: 17–25.
- Sahar AT, Zakia MA, Sobieh ShS. Assessment of Punica granatum L. extract on the mitotic arrest of plant bioassay system. Life Sci J., 2014; 11(8):757-770.
- Sahar M. Ouda, Antifungal Activity of Silver and Copper Nanoparticles on Two Plant Pathogens, Alternaria alternataand Botrytis cinerea. Research Journal of Microbiology, 2014; 9: 34-42.
- Sheikpranbabu, S., Kalishwaralal, K., Lee, K., The inhibition of advanced glycation end-products- induced retinal vascular permeability by silver nanoparticles. Biomaterials, 2010; 31: 2260–2271.
- Shunmugadevi C and PalanisamyP. Comparative Study of Silver Nanoparticles: Green Synthesis, Characterization and Biological Evaluation of Carica papaya and Andrographis paniculata Leaf Extract. World J Pharm Sci., 2016 ; 4(6): 454-465.
- Siddhanta, S., Zheng, C., Narayana, C., and Barman, I. An impediment to random walk: trehalose microenvironment drives preferential endocytic uptake of plasmonic nanoparticles. Chem. Sci., 2016; 7: 3730–3736.
- Sobhy I. I. Abdel-Hafez; Nivien A. Nafady; Ismail R. Abdel-Rahim; Abeer M. Shaltout; Jose´-Antonio Daro‘s ; Mohamed A. Mohamed 2016. Assessment of protein silver nanoparticles toxicity against pathogenic Alternaria solani 3 Biotech 6(199): 1-12.
- Supriyo C., Arpita B., and Surekha K. Green synthesis of protein capped silver nanoparticles from phytopathogenic fungus Macrophomina phaseolina(Tassi) Goid with antimicrobial properties against multidrug-resistant bacteria. Nanoscale Res Lett., 2014; 9(1): 365.1-11.
- Vahdati AR, Sadeghi B. A study on the assessment of DNA strand-breaking activity by silver and silica nanoparticles. J Nanostruct Chem., 2013; 1:1–3.
- Wu D, Fan W, Kishen A, Gutmann JL, Fan B. Evaluation of the antibacterial efficacy of silver nanoparticles against Enterococcus faecalis biofilm. J Endod, 2014; 40:285-290.
- Yu S-j, Yin Y-g, Liu J-f. Silver nanoparticles in the environment. Environ Sci Proc Impacts, 2013; 15:78-92.
- Zhi-Kuan Xia, Qiu-Hua Ma, Shu-Yi Li , De-Quan Zhang , Lin Cong , Yan-Li Tian , Rong-Ya Yang. The antifungal effect of silver nanoparticles on Trichosporon asahii . Journal of Microbiology, Immunology and Infection, 2016; 49: 182-188.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


