ISSN: 0973-7510
E-ISSN: 2581-690X
In the last two decades, there has been a remarkable rise in the instances of nosocomial infections associated with antibiotic-resistant Acinetobacter baumannii. The aimed of this study to determine the antibiotic resistance patterns and genetic relationship among isolates of carbapenem-resistant A. baumannii (CRAB) isolated from the clinical specimens of various inpatients. A total of sixty clinical isolates of A. baumannii were collected during February to September 2014 from King Fahd Hospital of the University (KFHU) in Khobar, Saudi Arabia. Antimicrobial susceptibility was tested using the Vitek 2 system and the minimum inhibitory concentrations (MICs) were estimated following the guidelines of the Clinical and Laboratory Standards Institute. Molecular epidemiological analysis of carbapenem-resistant A. baumannii (CRAB) was carried out by using enterobacterial repetitive intergenic consensus (ERIC-PCR). All of tested clinical isolates of A. baumannii in this study were associated with nosocomial infections and the highest rates of A. baumannii isolates were from wound swabs (23.3%) and transtracheal aspiration (20%). All of the 60 analyzed strains of A. baumannii in this study were classified as extensively drug-resistant (XDR), and the rates of antibiotic resistance against imipenem and meropenem were 93.3% and 96.6% respectively, while just 6.6% of strains were resistant to tigecycline. All 60 XDR A. baumannii isolates were sensitive only to colistin. The genotypic analysis of CRAB was performed by enterobacterial repetitive intergenic consensus (ERIC-PCR). ERIC-PCR able to discriminate the CRAB strains into four distinct clusters (A, B, C, and D) with genetic similarity ranged from 82.5 to 100%.
Carbapenem, Extensively Drug Resistant, Enterobacterial Repetitive Intergenic Consensus, ERIC-PCR, Saudi Arabia, Acientobacter baumannii
Acinetobacter baumannii is an encapsulated, catalase-positive, non-motile, Gram-negative coccobacilli bacterium1. This species is widely distributed in the environment and can be found in water, soil, and, occasionally, in human skin and throat as a normal microbiota. The presence of this species in the environment suggests an ability to persist desiccation and survive in the environment for long periods2. This phenomenon of adverse environmental persistence has increased the instance of hospital infections. Moreover, A. baumannii has the ability to adhere to hospital equipment, patients’ skin, and on the hands of doctors and nurses3,4. This species is responsible for numerous infections associated with skin, bloodstream, burns patients, and respiratory and urinary tract infections1,5. The use of mechanical ventilation and burn patients admitted to intensive care units (ICUs) are risk factors for acquiring infection with this species6,7. A. baumannii is considered among the most flourishing bacterial pathogens in acquiring antibiotic-resistant genes and has developed resistance to most of the available therapeutic antibiotics8. This species has multiple antibiotic resistance mechanisms, such as b-lactamases, aminoglycoside enzymes, altered target sites, efflux pump mechanisms, and permeability defects. Therefore, these mechanisms play a significant role in decreasing the therapeutic action of available antibiotics for the treatment of infections associated with A. baumannii9,10.
Multi-resistant, extensive-resistant, and pan-drug resistant (MDR, XDR, and PDR) A. baumannii strains are on the rise worldwide and present infection control and treatment challenges for clinicians and clinical microbiologists11-13. The evolution and spread of A. baumannii resistant to most of the available antimicrobial agents pose problems for future management since the pathogen plays a role in nosocomial infections. Currently, the management of MDR, XDR, and PDR A. baumannii infections poses serious clinical and epidemiological challenges14. The problem of antimicrobial resistance is of international concern because A. baumannii strains can display resistance to most available antibiotics, making treatment of these infections complicated15. The Centers for Diseases Control and Prevention (CDC) has classified MDR A. baumannii as a serious threat pathogen9. This species is also categorized by the Infectious Diseases Society of America (IDSA) as a superbug and is one of the six significant “ESKAPE” pathogens, which include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species “ESKAPE”16. The current study aimed to determine the antibiotic resistance patterns in isolates of A. baumannii from various inpatients’ clinical specimens and to determine the genetic relationship among the isolates of carbapenem-resistant A. baumannii (CRAB) isolated in King Fahd Hospital of the University (KFHU) from February to September 2014 in Khobar, Saudi Arabia.
Ethics Statement
The ethics committee of the Imam Abdulrahman Bin Faisal University approved this study (IRB -2017-01-203).
Bacterial Isolates
From February to September 2014, a total of 60 A. baumannii clinical isolates were analyzed from a 450-bed teaching hospital. The selection criteria for isolates were only carbapenem-resistance and meeting the definition of XDR strains using the definition criteria described by Magiorakos et al. (2012)12. All hospital isolates of A. baumannii were identified by the Vitek 2 automatic system and further confirmed using the API 20 NE system (BioMe´rieux, France).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was tested using the Vitek 2 system (BioMérieux, France) and the minimum inhibitory concentrations (MICs) were estimated following the guidelines of the Clinical and Laboratory Standards Institute (17). The antibiotics tested in this study were ampicillin/sulbactam (AM/SUL), amikacin (AK), cefepime (FEP), ceftazidime (CAZ), ciprofloxacin (CIP), colistin (CL), gentamicin (GM), imipenem (IMP), meropenem (MEP), piperacillin/tazobactam (PIP/TAZ), tobramycin (TOB) and tigecycline (TIG).
DNA Extraction
Isolates of A. baumannii were grown in Luria Bertani (LB) broth and incubated in a thermal shaker at 37°C for 24 h. Incubated LB broth (1.5 ml) was then harvested and centrifuged for 1 min at 10,000 rpm, and the obtained pellet was suspended in 700 µl of deionized water and boiled at 100°C for 15 min to lyse the cells and free the DNA. The extracted DNA for all isolates was immediately stored at -20°C and used as a template for ERIC-PCR.
Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction
The genetic relationship between 56 carbapenem-resistant isolates was determined using the enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) method as described elsewhere18.
ERIC-PCR Data Analysis
The DNA polymorphism patterns were obtained by electrophoresis separation and were analyzed using GelJ software19. The DNA fingerprints for all gel images were calculated using the Dice coefficient. The unweighted pair-group method using arithmetic averages (UPGMAs) was used to construct the phylogenetic tree for CRAB isolates. The clonal relationship was constructed based on the similarity matrix, and a dendrogram was generated by ERIC fingerprints among the isolates. Isolates in the clustered dendrogram exceeding 85% similarity are were considered clonally related.
Statistical analysis
The Chi-square tests and other analyses were carried out using MS Excel 2007 (Microsoft, Redmond, WA, USA) and PAST statistical software (version 2.04), as described by Hammer20.
A total of 60 isolates of A. baumannii were isolated from various clinical specimens of inpatients admitted to the KFHU (Table 1). The highest rates of A. baumannii isolation were associated with wound swab (23.3%) and transtracheal (20%) specimens. According to the chi-square goodness-of-fit test, there was no association between gender and specimen (P = 0.22). Most of the A. baumannii strains were isolated from adult patients (Table 2).
Table (1):
The distribution of A. baumannii in clinical specimens.
Specimen |
Male |
Female |
Total |
|---|---|---|---|
Abscess |
0 |
1 |
1 |
Blood |
4 |
0 |
4 |
Intravenous catheter tip |
3 |
0 |
3 |
Nasal Swab |
1 |
0 |
1 |
Peritoneal fluid |
2 |
0 |
2 |
Pleural fluid |
1 |
1 |
2 |
Rectal swab |
3 |
3 |
6 |
Skin swab |
1 |
0 |
1 |
Sputum |
2 |
2 |
4 |
Throat swab |
4 |
1 |
5 |
Transtracheal aspiration |
7 |
5 |
12 |
Urine |
5 |
0 |
5 |
Wound swab |
6 |
8 |
14 |
Total |
39 |
21 |
60 |
There was no association between gender and specimen (p = 0.22).
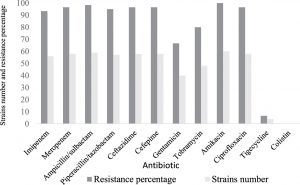
All of the analyzed strains of A. baumannii in this study were classified as XDR, and no PDR isolate was detected (Table 3). The percentage of antibiotic resistance and the MICs of the A. baumannii are shown in (Fig. 1) and (Table 3). Evaluating all of the major classes of antibiotics, we found that all of the isolates were susceptible to colistin (Table 3). However, 93.3% and 96.6% of strains were resistant to imipenem and meropenem respectively (Table 3). Just 6.6% of strains were resistant to tigecycline (Fig. 1) (Table 3). All the study isolates were resistant to amikacin, with similar MIC values (all > 16 µg/mL) (Table 3).
Table (2):
The distribution of A. baumannii strains in different age groups.
Specimen |
0–25 years |
26–55 years |
56 and above |
|---|---|---|---|
Abscess |
0 |
1 |
0 |
Blood |
0 |
3 |
1 |
Intravenous catheter tip |
0 |
0 |
3 |
Nasal Swab |
0 |
1 |
0 |
Peritoneal fluid |
0 |
1 |
1 |
Pleural fluid |
0 |
0 |
2 |
Rectal swab |
1 |
4 |
1 |
Skin swab |
0 |
0 |
1 |
Sputum |
0 |
2 |
2 |
Throat swab |
1 |
3 |
1 |
Transtracheal aspiration |
0 |
3 |
9 |
Urine |
0 |
1 |
4 |
Wound swab |
2 |
6 |
6 |
Total (60) |
4 |
25 |
31 |
There was no association between months and drug (p = 0.537).
Table (3):
Minimum inhibitory concentrations breakpoint for an extensively drug-resistant strain of A. baumannii.
| Antibiotic | MIC breakpoint (µg/mL) | ||
|---|---|---|---|
| < 1 | Between 2 and 8 | > 16 | |
| Imipenem | 4 | 0 | 56 |
| Meropenem | 2 | 0 | 58 |
| Ampicillin/sulbactam | 0 | 5 | 55 |
| Piperacillin/tazobactam | 0 | 2 | 58 |
| Ceftazidime | 0 | 2 | 58 |
| Cefepime | 0 | 2 | 58 |
| Gentamicin | 15 | 5 | 40 |
| Tobramycin | 10 | 2 | 48 |
| Amikacin | 0 | 0 | 60 |
| Ciprofloxacin | 2 | 58 | 0 |
| Tigecycline | 18 | 42 | 0 |
| Colistin | 60 | 0 | 0 |
There was no association between MIC and drug (p < 0.001).
Overall, the highest rates of nosocomial XDR of A. baumannii isolates at our hospital were reported between Februarys to September 2014, while the highest number of CRAB isolates were reported between May to July 2014 (Table 4). According to the chi-square goodness-of-fit test, there was no association between month of isolation and drug resistance (p = 0.999).
Table (4):
The resistance rates of nosocomial A. baumannii strains between February and
September 2014.
| Drugs | Months (n) | ||
|---|---|---|---|
| Feb-March (6) | May-July (42) | Aug-Sept (12) | |
| Imipenem | 5 | 40 | 11 |
| Meropenem | 6 | 41 | 11 |
| Ampicillin/sulbactam | 6 | 42 | 11 |
| Piperacillin/tazobactam | 6 | 41 | 10 |
| Ceftazidime | 6 | 41 | 11 |
| Cefepime | 6 | 41 | 11 |
| Gentamicin | 4 | 29 | 7 |
| Tobramycin | 5 | 36 | 7 |
| Amikacin | 6 | 42 | 12 |
| Ciprofloxacin | 6 | 41 | 11 |
| Tigecycline | 1 | 2 | 2 |
| Colistin | 0 | 0 | 0 |
There was no association between the months and drug (p = 0.999).
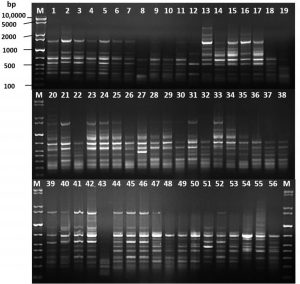
Fig. 2. DNA fingerprint analysis of the 56 carbapenem-resistant Acinetobacter baumannii by ERIC-PCR. M: GelPilot 1 Kb Plus ladder
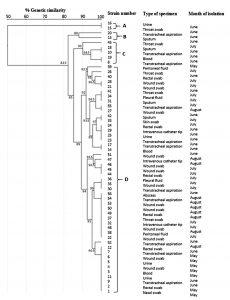
The ERIC-PCR technique was used for the molecular typing of 56 CRAB strains. The banding patterns consisted of 5 to 14 fragments per strain. The apparent molecular sizes of the band fragments ranged from 200 to 3,900 bp (Fig. 2). The ERIC dendrogram revealed four distinct clusters (A, B, C, and D) with genetic similarity ranging from 82.5 to 100% (Fig. 3). These clustered strains were recovered from clinical inpatients specimens throughout the four months (May, June, July, and August), raising concerns about the potential persistence of the epidemic clonal strain. Among the four clusters, Cluster D accounted for 47 (84%) of the isolates, suggesting a high genetic relatedness among the CRAB genotype carrying strains (Fig. 3).
Fig. 3. Dendrogram based on clusters analysis of ERIC-PCR fingerprint data illustrating the relationships among the carbapenem-resistant Acinetobacter baumannii strains. The genetic similarities between the fingerprints were calculated using the Dice coefficient (optimization, 1%; position tolerance, 1%), and the fingerprints were grouped according to their similarities using the unweighted pair-group method using arithmetic averages method
The strains of Cluster D were highly genetically similar, raising the possibility of cross-transmission within some of the hospital wards, including ICU wards. The other three minor clusters (A, B, and C) account for 16% of the recovered isolates. Our findings demonstrate that the ERIC-PCR method is a rapid, simple typing technique with a level of discrimination equivalent to that of pulsed-field gel electrophoresis (PFGE).
A. baumannii has been implicated in many types of nosocomial infection in healthcare settings19. Recently, the number of studies documenting epidemics of CRAB outbreaks in hospital ICUs has increased21,22. An increased rate has been reported in Saudi hospital ICUs, particularly among immunocompromised patients23,24. In this study, all of the A. baumannii isolates associated with inpatient infections were classified and identified as XDR (Table 3). The overall resistance rate of A. baumannii to the tested antibiotics was 84.2%. The antibiotic susceptibility testing revealed that 56 (93.3%) A. baumannii isolates were carbapenem-resistant. Thus, this reported rate of resistance is similar to those reported in other countries, as discussed elsewhere21,22. Our data is in line with other studies conducted in Saudi Arabia, and the Gulf Cooperation Council States have reported a significantly increased rated of CRAB isolates23-25. We detected four strains (6.6%) resistant to tigecycline (Fig. 1). Several studies have reported an increased resistant to tigecycline, which has created a therapeutic challengefor clinicians, as discussed elsewhere26-28.
DNA-based molecular epidemiology typing techniques are important tools to investigate and track outbreaks of bacterial strains and to the control the spread of bacterial isolates associated with nosocomial infections in healthcare settings29. In this study, we used ERIC-PCR to determine the genetic relatedness of 56 CRAB isolates. By this approach, we detected an increased rate of detection of CRAB isolates between May and August 2014 (Table 4). Interestingly, based on dendrogram cluster analysis of ERIC-PCR fingerprint data, a single genetically-related cluster (Cluster D) was shown to account for 47 of the 56 (84%) CRAB isolates. This is likely due to the spread of clonally related CRAB isolates, which were responsible for nosocomial infections between May and August 2014. Molecular typing can be used as an epidemiological method to type bacterial strains and thus monitor and track the spread of major nosocomial infections pathogens30. Rapid molecular typing methods have become popular in clinical microbiology research laboratories and play a significant role in decontamination and controlling the spread of infection in hospitals30. The inclusion of molecular typing techniques to determinate microbial clonality as part of routine infection control programs within healthcare settings will be of medical and economic benefit31.
Here we detected a rapid increase in the isolation of XDR A. baumannii and CRAB infections at our hospital between May and August 2014. ERIC-PCR was shown to be useful for typing XDR A. baumannii and CRAB, thus providing tools for epidemiological and clinical follow-up studies. The isolation of similar clones (> 90% genetic similarity) from different specimens within a single hospital suggests horizontal transmission, and it is possible that these related isolates of XDR A. baumannii and CRAB belonged to a single bacterial clone circulating through the hospital setting. The frequency of isolation of these drug-resistant bacteria is a reminder of the importance of stewardship surveillance for resistant bacteria in the hospital settings to prevent their spread and epidemy, and the importance of strict implementation of infection control guidelines to prevent circulation through the hospital setting. This study established baseline evidence of clonal dissemination of closely related CRAB at our hospital, indicating the need for further surveillance.
Acknowledgements
The authors would like to thank the laboratory technicians in clinical microbiology laboratory of King Fahd Hospital of the University (KFHU) for samples collection during the study period. This study was supported by the Deanship of Scientific Research at the Imam Abdulrahman Bin Faisal University, grant No. 2013203.
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538-82. doi: 10.1128/CMR.00058-07. [PubMed: 18625687].
- Hrenovic J, Durn G, Goic-Barisic I, Kovacic A. Occurrence of an environmental Acinetobacter baumannii strain similar to a clinical isolate in paleosol from Croatia. Appl Environ Microbiol. 2014;80(9):2860-6. doi: 10.1128/AEM.00312-14. [PubMed: 24584245].
- de Abreu PM, Farias PG, Paiva GS, Almeida AM, Morais PV. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: a potential health hazard. BMC Microbiol. 2014;14(1):118. doi: 10.1186/1471-2180-14-118. [PubMed:24885173].
- Greene C, Wu J, Rickard AH, Xi C. Evaluation of the ability of Acinetobacter baumannii to form biofilms on six different biomedical relevant surfaces. Lett Appl Microbiol. 2016;63(4):233-9. doi: 10.1111/lam.12627. [PubMed: 27479925].
- Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence. 2012;3(3):243-50. doi: 10.4161/viru.19700. [PubMed: 22546906].
- Falagas M, Karveli E, Siempos I, Vardakas K. Acinetobacter infections: a growing threat for critically ill patients. Epidemiol Infect. 2008;136(8):1009-19. doi: 10.1017/S0950268807009478. [PubMed: 17892629].
- Alsan M, Klompas M. Acinetobacter baumannii: an emerging and important pathogen. J Clin Outcomes Manag. 2010;17(8):363. [PubMed: 26966345].
- Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [PubMed: 28348979].
- Clark NM, Zhanel GG, Lynch III JP. Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr Opin Crit Care. 2016;22(5):491-9. doi: 10.1097/MCC.0000000000000337. [PubMed: 27552304].
- Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol spectr. 2016;4(2). [PubMed: 27227291].
- MacDougall C, Polk RE. Antimicrobial stewardship programs in health care systems. Clin Microbiol Rev. 2005;18(4):638-56. doi: 10.1128/CMR.18.4.638-656.2005. [PubMed: 16223951].
- Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug resistant, extensively drug resistant and pandrug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-81. doi: 10.1111/j.1469-0691.2011.03570.x. [PubMed: 21793988].
- Manchanda V, Sanchaita S, Singh N. Multidrug resistant Acinetobacter. J Glob Infect Dis. 2010;2(3):291. doi: 10.4103/0974-777X.68538. [PubMed: 20927292].
- Swe-Han KS, Mlisana KP, Pillay M. Analysis of clinical and microbiological data on Acinetobacter baumannii strains assist the preauthorization of antibiotics at the patient level for an effective antibiotic stewardship program. J Infect Public Health. 2017;10(5):608-16. doi: 10.1016/j.jiph.2017.01.014. [PubMed: 28237694].
- Hawkey P. The growing burden of antimicrobial resistance. J Antimicrob Chemother. 2008; 62(suppl_1):i1-i9. doi: 0.1093/jac/dkn241. [PubMed: 18684701].
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1-12. doi: 10.1086/595011. [PubMed: 19035777].
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI Document M100-S21. Wayne, PA: CLSI; 2011.
- Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991;19(24):6823-31. [PubMed: 1762913].
- Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, et al. GelJ–a tool for analyzing DNA fingerprint gel images. BMC bioinformatics. 2015;16(1):270. doi: 10.1186/s12859-015-0703-0. [PubMed: 26307353].
- Hammer X, Harper D, Ryan P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron. 4: 9pp. 2001.
- Hammoudi D, Moubareck CA, Hakime N, Houmani M, Barakat A, Najjar Z, et al. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int J Infect Dis. 2015;36:56-61. doi: 10.1016/j.ijid.2015.05.015. [PubMed: 26004171].
- Bianco A, Quirino A, Giordano M, Marano V, Rizzo C, Liberto MC, et al. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect Dis. 2016;16(1):747. doi: 10.1186/s12879-016-2036-7. [PubMed: 27955639].
- Alsultan AA, Aboulmagd E, Evans BA, Amyes SG. Clonal diversity of Acinetobacter baumannii from diabetic patients in Saudi Arabian hospitals. J Med Microbiol. 2014;63(11):1460-6. doi: 10.1099/jmm.0.079640-0. [PubMed: 25106863].
- Al-Obeid S, Jabri L, Al-Agamy M, Al-Omari A, Shibl A. Epidemiology of extensive drug resistant Acinetobacter baumannii (XDRAB) at Security Forces Hospital (SFH) in Kingdom of Saudi Arabia (KSA). Journal of Chemotherapy. 2015;27(3):156-62. doi: 10.1179/1973947815Y.0000000019. [PubMed: 25867622].
- Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, et al. Molecular Epidemiology of carbapenem resistant Acinetobacter baumannii in the Gulf Cooperation Council States. Dominance of OXA-23-type producers. J. Clin Microbiol. 2015:JCM. 02784-14. doi: 10.1128/JCM.02784-14. [PubMed: 25568439].
- Navon-Venezia S, Leavitt A, Carmeli Y. High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2007;59(4):772-4. doi: 10.1093/jac/dkm018. [PubMed: 17353223].
- Baadani AM, Thawadi SI, El-Khizzi NA, Omrani AS. Prevalence of colistin and tigecycline resistance in Acinetobacter baumannii clinical isolates from 2 hospitals in Riyadh Region over a 2-year period. Saudi Med J. 2013;34(3):248-53. [PubMed: 23475088].
- Deng M, Zhu M-H, Li J-J, Bi S, Sheng Z-K, Hu F-S, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014;58(1):297-303. doi: 10.1128/AAC.01727-13. [PubMed: 24165187].
- Ranjbar R, Karami A, Farshad S, Giammanco G, Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol. 2014;37(1):1-15. [PubMed: 24531166].
- Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43(1):199-207. doi: 10.1128/JCM.43.1.199-207.2005. [PubMed: 15634972].
- Hacek DM, Suriano T, Noskin GA, Kruszynski J, Reisberg B, Peterson LR. Medical and economic benefit of a comprehensive infection control program that includes routine determination of microbial clonality. Am J Clin Pathol. 1999;111(5):647-54. [PubMed: 10230355].
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.