ISSN: 0973-7510
E-ISSN: 2581-690X
Infection with the hepatitis C virus (HCV) is serious and may lead to death. Venous blood samples were collected from HCV-infected patients (n=60) and from people without infection (n=60) as controls. These patients visited the Digestive System and Liver Disease Hospital, Baghdad, Iraq, during the period from 1st February to 1st November 2021. The results revealed no significant difference between males (31; 50.8%) and females(29; 49.2%) HCV-infected patients. The highest infection rate (55.6%) was observed among the 50–78 years age group, then 53.7% among the 25–49 years age group, then 36.7% among the 5–24 years age group with a significant difference (P=0.2). A highly significant difference was observed in the mean glutamic-pyruvic transaminase levels between HCV-infected patients (25.56±12.45) and the controls (17.86±4.91) (P=0.01), and a significant difference was observed in the mean glutamic-oxaloacetic transaminase levels between HCV-infected patients (21.70±8.63) and the controls (15.93±4.35) (P=0.02). A highly significant difference was demonstrated in mean alkaline phosphatase levels between HCV-infected patients (362.15±113.60) and controls (197.81±34.70)(P=0.001). Overall, we found that blood markers could aid in disease diagnosis and prognosis rather than mutations within the nonstructural 5A.1 hot spot. The frequency of mutations within this site was found to be very low.
Sequence, Hepatitis C Virus (HCV), Liver diseases
Infection with the Hepatitis C virus (HCV) is dangerous and may be fatal. There are three types of infections: acute hepatitis, which is short-lived, lasts for 6 months, and then the body recovers from it.1 Chronic hepatitis C occurs for a longer period of more than 6 months (approximately 85% of the cases) and is equal to infection with hepatitis C, which may lead to health problems such as cirrhosis and liver cancer.2,3 In cirrhosis, fibrosis leads to the replacement of healthy hepatic parenchyma cells with scar tissue4; however, it occurs over a prolonged period from 20 to 30 years or may be faster when the patient consumes alcohol and suffers from acquired immunodeficiency.1,5
Liver enzymes are commonly used to evaluate the pathology of various diseases, including viruses such as HCV.6 The scale of these enzymes is classically used to provide information about viral hepatitis infections if the main disorder is biliary hepatitis, and knowing the percentage of enzymes in the sera of infected people, as well as knowing the patterns, provides abundant information for diagnosing the pathological condition.7,8 There are multiple blood abnormalities accompanying chronic viral hepatitis, such as iron deficiency anemia,9 which are frequent complications of chronic liver diseases. These are caused by many factors, including chronic bleeding in the digestive system.10,11 and the diagnosis of anemia caused by iron deficiency is very difficult, as the diagnosis of iron, ferritin, and transferrin saturation in the average body size with the liver disease itself, leads to difficulty in interpreting the results.12,13
Molecular studies of HCV infections and the pathophysiology of infections lead to immune liver damage, cirrhosis, and lipid metabolism.14,15 For amplification of the antigenic site NS3.1 with nonstructural 5A (NS5A). 1, specific primers with restriction enzymesites16 have been designed. For the above primers, Hind-III with Xho-I restriction enzyme site was added to primers, whereas primers of NS5A-1 contained EcoR-1 with Hind-III restriction site.17 HCV combination therapies with mutations at NS5-A-PKRBD and NS5A-ISDR with NS5A-V3 HCV regions were made. There was also a change from isoleucine to valine(I2252 V).18,19 Hepatitis C virus is a risky infection which may lead to death20,21 In addition, the baseline NS5A RASs finger print was assessed in 185-DAA treatment-naive GT-1b patients using next-generation sequencing methods. Patients who presented with Y93-H had higher HCV NS5-sequences entropy. Our study aimed to detect mutations in the Ns5atp4 NS5A (hepatitis C virus) gene in HCV infections.
Venous blood samples were collected from 60 HCV-infected patients and 60 patients without infection as controls. The patients visited the Digestive System and Liver Disease Hospitals in Baghdad, Iraq, from 1st February to November 1, 2021. Serum levels of glutamic-pyruvic transaminase (GPT), glutamic-oxaloacetic transaminase (GOT), and alkaline phosphatase (ALP) were determined using a fully automatic analyzer (Cobas Integra 400 Roche, Germany), and gene sequences were further investigated by PCR and Sanger sequencing (Thermo Fisher Scientific) in the above hepatitis patients to detect the Ns5atp4 NS5A (hepatitis C virus) gene. The primers used were: forward 52 – GAGGAAGTGCTTAGGGCGTT 32 -, reverse: 52 – CATCCGTGCCTCTGAGATCC32 -, with a product length of 379.
DNA extraction
Within 60 min of sample collection, centrifugation was done at 1600 ׳ g for 20 min to separate plasma from peripheral blood cells from blood collected in EDTA tubes. Blood plasma was frozen at 80°C in RNA later TM (AM7021) until DNA could be extracted. A circulating nucleic acid kit (Thermo Fisher Scientific) was used to extract DNA from plasma samples (2 ml), following the manufacturer’s protocol.
Quantity and Quality Determination for DNA
A NanoDrop Lite UV Spectrophotometer (Thermo Scientific™) was used to assess the purity of DNA, estimated by reading the optical density at A260. DNA quality was estimated by measuring the optical density at 280nm, whereas the ratio of optical densities was calculated at 260/280 nm.
Statistical analyses
Data were analyzed using the SPSS version-20 program. Student’s t-test was used to detect the variation between groups.22 P<0.05 was considered statistically significant.
According to sex distributions among the study groups, there was no significant difference between males(31; 50.8%) and females (29; 49.2%), as illustrated in Table 1.
Table (1):
Distribution of the study groups by gender.
| Gender | Study groups | ||
|---|---|---|---|
| Patient | Control | ||
| Males | N | 31 | 30 |
| % | 50.8% | 49.2% | |
| Females | N | 29 | 30 |
| % | 49.2% | 50.8% | |
| Total | N | 60 | 60 |
| % | 100% | 100% | |
| Chi square | 0.03 | ||
| P value | 0.8 (N.S) | ||
Table 2 shows that the highest infections rates (55.6%) was among the group (50-78) year, followed by 53.7% in the group25-49 year, then 36.7% in the group 5-24 year, (P=0.2).
Table (2):
Prevalence of the study groups according to age groups.
| Age group (years) | Study group | |
|---|---|---|
| Patient | Controls | |
| (5-24) | 11 | 19 |
| 36.7% | 63.3% | |
| (25-49) | 29 | 25 |
| 53.7% | 46.3% | |
| (50-78) | 20 | 16 |
| 55.6% | 44.4% | |
| Total | 60 | 60 |
| 100% | 100% | |
| Chi square | 2.87 | |
| P value | 0.2 | |
Table 3 showed that a highly significant difference was demonstrated in the mean GPT levels between HCV-infected patients (25.56±12.45) and the controls (17.86±4.91) (P = 0.01), and a significant difference was demonstrated in the mean GOT levels between HCV-infected patients (21.70±8.63) and the controls (15.93±4.35) (P = 0.02). A highly significant difference was demonstrated in mean ALP levels between HCV-infected patients (362.15±113.60) and controls (197.81±34.70) (P = 0.001).
Table (3):
Distribution of mean serum levels of GPT, GOT and ALP among the study groups.
Parameter |
Groups |
N |
Means |
T test |
P values |
|---|---|---|---|---|---|
GPT (0-60) |
Patients |
60 |
25.56±12.45 |
2.43 |
0.01 |
Control |
60 |
17.86±4.91 |
|||
GOT (0-50) |
Patient |
60 |
21.70±8.63 |
2.33 |
0.02 |
Control |
60 |
15.93±4.35 |
|||
ALP (80-360) |
Patient |
60 |
362.15±113.60 |
6.87 |
0.001 |
Control |
60 |
197.81±34.70 |
The PCR products were observed to be approximately 359bp (Figure 1), as can be seen from the agarose gel (1.2%).In the genetic sequence of HCV, there were mutations in the Ns5atp4 NS5-A (hepatitis C virus) gene at several locations, which increases virulence severity. In lane (1) reference the Rattus norvegicus gene, the change occurred in lane 2 Rattus norvegicus, C>A, T>C, T>G, G>A, G>A, A>C, G>C, G>T, C>T, C>T, C>T, G>C, T>A, G>T, T>A, C>T, G>C, C>G, C>A, C>A, C>T, A>T as shown in Figure 2.
Figure 1. 1.2%agarose gel showing the amplified bands of genotypes of Ns5atp4 NS5A (hepatitis C virus). PCR products can be seen approximately at 379bp. Lane M: molecular marker (100bp). Lane 1: reference gene; Lane 2–10: samples
Figure 2. Changes in the lane2 Rattusnorvegicus gene, C>A, T>C, T>G, G>A, G>A, A>C, G>C,G>T, C>T, C>T, C>T, G>C, T>A, G>T, T>A, C>T, G>C, C>G, C>A, C>A, C>T, A>T
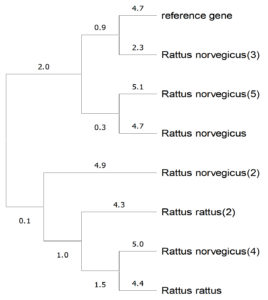
Figure 3. Phylogenic tree of the Rattus norvegicus gene isolated from Iraqi hepatitis C virus-infected patients
From the phylogenetic tree (Figure 3), we observed that the sample sequences were a close match to the reference gene, but with base pair alterations, as seen from the node lengths. These mutations need to be traced further to determine whether they belong to point or silent mutations.
Infection with HCV is dangerous and may be fatal. The results of our study showed no significant differences according to sex among the HCV-infected patients. These findings agreed with those of a previous study,23 which reported that 434 (56%) of patients were male and 341 (44%) were female.23 The highest infection rate (55.6%) was detected in the 50–78 years age group compared to that in other age groups, and these results disagreed with those of Gheyath et al., 2018, who reported that the infection in younger patients was the highest in comparison with that in other ages.24 The results of the mean serum GPT levels found in our study among the study groups matched with those of Fabrizi et al.24 who reported that the highest elevation in GPT levels was observed in HCV-infected patients because the infection may cause hepatocyte damage leading to the exit of the enzyme in high concentrations.25 Wanachiwanawin (2003) reported similar results; when compared to patients without anti-HCV antibodies, patients with anti-HCV antibodies had substantially abnormal liver functions, including lower levels of serum albumin and higher levels of aspartate aminotransferase (SGOT) and alanine aminotransferase (SGPT) (P<0.017).26
Serum ALP levels can detect significant fibrosis (S2) in HBeAg(+) HCV patients who have not received therapy, which may lessen the need for liver biopsies and aid in direct clinical CHB treatment.27,28 The mean serum ALP level in HCV-infected patients was found to be 362.15±113.60, whereas in the controls it was 197.81±34.70. Hu Jianhua et al.26 stated that there is a direct relationship between viral hepatitis and ALP levels, where there is an obvious increase in the enzyme levels, although it is frequently excreted with viral liver diseases.29 Bodlaj et al. reported from their findings that following therapy, ALP levels reduced to the normal range from abnormal levels. Hu et al.26 reported that healthy controls had ALP levels that were significantly lower than those of CHB patients. Additionally, ALP gradually increased in all patients and separately in HBeAg(-) HCV patients with the progression of liver fibrosis, but not in HBeAg(+) HCV patients.29,30
Infection with HCV is one of the main causes of chronic liver disease worldwide. Response to interferon therapy may be correlated with mutations in various HCV subtype 1b (HCV-1b) NS5A gene regions.31,32 Direct-acting antivirals (DAAs) are an efficient means of treating HCV, which places a significant burden on health. One of the targets of DAAs is NS5A, which is involved in the propagation of viral genomes.33,34
The virus was found in several locations, which increased its virulence severity, with a change in the Rattus norvegicus gene. Zhou et al. (2021)34 stated that there were frequent genetic mutations in the Ns5atp4 NS5A (hepatitis C virus) gene that increased the virulence of the virus, which may lead to hepatocyte destruction, liver cirrhosis, or even liver cancer, which may be fatal in most cases.35 Rashed et al. (2021) affirmed that mutations in HCV are common, and this genetic change is what makes this epidemic virus remain for a long time in the liver of the infected people to be able to destroy it.36 Mejer et al. (2020) reported that mutations in this gene make the virus more resistant to treatments.36-38
HCV infection affects the health of the general public. It is challenging to identify polymorphisms and variants that contain substitutions linked to resilience to novel antivirals because of the high rate of HCV replication and the absence of post-transcriptional correction mechanisms. In our study, we found elevated levels of GPT and ALP in the diseased patients. We also found one single mutation within the Ns5atp4 NS5A region, which is a very common hotspot for mutations. However, the frequency of mutation is not seen in other samples, which suggests that a mutation study within this region might not help in diagnosis or prognosis. Rather, blood markers would be more reliable in disease diagnosis.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Jaleel Najah Samanje’s invaluable assistance and for providing all feasible statistical analysis facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by Iraqi Ministry of Health from 1st February to 1st November 2022 under no. 2380.
- Ratini M. Hepatitis C and the Hep C Virus. Web Med. 2021;23-25.
- Wing PAC, Jones M, Cheung M, et al. Amino acid substitutions in genotype 3a hepatitis C virus polymerase protein affect responses to sofosbuvir. Gastroenterology. 2019;157(3):692-704.
Crossref - Suda G, Kimura M, Shigesawa T, et al. Effects of resistance-associated variants in genotype 2 hepatitis C virus on viral replication and susceptibility to antihepatitis C virus drugs. Hepatol Res. 2019;49(11):1275-1285.
Crossref - Elbanan WK, Fathy SA, Ibrahim Hegazy MGA. Assessment of interleukin 17 and transforming growth factor-beta 1 in hepatitis C patients with disease progression. Trop Biomed. 2020;37(4):1093-1104.
Crossref - Kotsiri I, Hadziyannis E, Georgiou A, Papageorgiou MV, Vlachogiannakos I, Papatheodoridis G. Changes in serum transforming growth factor-ג1 levels in chronic hepatitis C patients under antiviral therapy. Ann Gastroenterol. 2016;29(1):79-84. PMID: 26752952
- Kliemann DA, Tovo CV, da Veiga AB, de Mattos AA, Wood C. Polymorphisms and resistance mutations of hepatitis C virus on sequences in the European hepatitis C virus database. World J Gastroenterol. 2016;22(40):8910-8917.
Crossref - Hall Ph, Cash J. What is the Real Function of the Liver ‘Function’Tests? Ulster Med J. 2012;81(1):30-36.
- Nakamura F, Takeda H, Ueda Y, et al. Mutational spectrum of hepatitis C virus in patients with chronic hepatitis C determined by single molecule real-time sequencing. Sci Rep. 2022;12(1):7083.
Crossref - Saif-Al-Islam M, Mohamed HS, Younis MA, Abdelhamid MY, Ali MM, Khalaf S. Impact of Gender Difference on Characteristics and Outcome of Chronic Hepatitis C. Open Journal of Gastroenterology: 2020;10(11):281-294.
Crossref - Gkamprela E., Deutsch M., Pectasides D. Iron deficiency anemia in chronic liver disease: etiopathogenesis, diagnosis and treatment, Ann Gastroenterol. 2017;30(4):405-413.
Crossref - Suan MAM, Said SM, Lim PY, Azman AZF, Hassan MRA. Risk factors for hepatitis C infection among adult patients in Kedah State, Malaysia. PLOS ONE. 2019;14(10):e0224459.
Crossref - Alexei V, Lagaye S. Hepatitis C virus: Morphogenesis, infection and therapy. World J Hepatol. 2018;10(2):186-212.
Crossref - Ramirez S, Bukh J. Current status and future development of infectious cell culture models for the major genotypes of hepatitis C virus: essential tools in testing of antivirals and emerging vaccine strategies. Antiviral Res. 2018;158:264-287.
Crossref - Sabri S, Khan MI, Rafique SH, Ali A, Khan MS. Identification and expression analysis of antigenic sites of hepatitis C virus genotype 3a NS3 and NS5A genes of local isolate. Egyptian Liver Journal. 2021;11(1):17.
Crossref - Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61(1):S45-S57.
Crossref - Poordad F, Dieterich D. Treating hepatitis C: current standard of care and emerging direct-acting antiviral agents. J Viral Hepat. 2012;19(7):449-464.
Crossref - Bokharaei-Salim F, Keyvani H, Salehi-Vaziri M, et al. Mutations in the NS5A gene of hepatitis C virus subtype 1b and response to peg-IFN a-2a/RBV combination therapy in Azerbaijani patients. Arch Virol. 2014;159(11):2893-2899.
Crossref - Lu J, Feng Y, Chen L, et al. Subtype-Specific Prevalence of Hepatitis C Virus NS5A Resistance Associated Substitutions in Mainland China. Front Microbiol. 2019.
Crossref - Tisthammer KH, Solis C, Orcales F, et al. Assessing in vivo mutation frequencies and creating a high-resolution genome-wide map of fitness costs of Hepatitis C virus. PLoS Genet. 2022;18(5):e1010179.
Crossref - Due OT, Thakkinstian A, Thavorncharoensap M, et al. Cost-utility analysis of direct-acting antivirals for treatment of chronic hepatitis C genotype 1 and 6 in Vietnam. Value in Health. 2020;23(9):1180-1190.
Crossref - Backus LI, Belperio PS, Shahoumian TA, Loomis TP, Mole LA. Real-world effectiveness and predictors of sustained virological response with all-oral therapy in 21, 242 hepatitis C genotype-1 patients. Antiv Ther. 2017;22(6):481-493.
Crossref - Welsch C, Efinger M, von Wagner M, Abu-raddad LJ. Ongoing liver inflammation in patients with chronic hepatitis C and sustained virological response. PLoS One. 2017;12(2):e0171755.
Crossref - Nasrallah GK, Dargham SR, Mohammed LI, et al. Estimating seroprevalence of herpes simplex virus type 1 among different Middle East and North African male populations residing in Qatar. J Med Virol. 2018;90(1):184-190.
Crossref - Fabrizi F, Cerutti R, Dixit V, Messa P. The impact of antiviral therapy for HCV on kidney disease: a systematic review and meta-analysis. Nefrologia. 2020;40(3):299-310.
Crossref - Wanachiwanawin W, Luengrojanakul P, Sirangkapracha P, Leowattana W, Fucharoen S. Prevalence and clinical significance of hepatitis C virus infection in Thai patients with thalassemia. Int J Hematol. 2003;78(4):374-378.
Crossref - Hu J, Zhang X, Gu J, et al. Serum alkaline phosphatase levels as a simple and useful test in screening for significant fibrosis in treatment-naive patients with hepatitis B e-antigen negative chronic hepatitis B. Eur J Gastroenterol Hepatol. 2019;31(7):817-823.
Crossref - Koenig G, Seneff S. Gamma-glutamyltransferase: a predictive biomarker of cellular antioxidant inadequacy and disease risk. Dis Markers. 2015;(4):818570.
Crossref - McDonald SA, Dahlui M, Mohamed R, Naning H, Shabaruddin FH, Kamarulzaman A. Projections of the current and future disease burden of hepatitis C virus infection in Malaysia. PLOS ONE. 2015;10(6):e0128091.
Crossref - Bodlaj G, Hubmann R, Saleh K, Stojakovic T, Biesenbach G, Berg J. Alkaline phosphatase predicts relapse in chronic hepatitis C patients with end-of-treatment response. World J Gastroenterol. 2010;16(19):2407-2710.
Crossref - Theys K, Feder AF, Gelbart M, Hartl M, Stern A, Pennings PS. Within-patient mutation frequencies reveal fitness costs of CpG dinucleotides and drastic amino acid changes in HIV. PLoS Genet. 2018;14:e1007420-24.
Crossref - Knops E, Sierra S, Kalaghatgi P, Heger E, Kaiser R, Kalinina OV. Epistatic Interactions in NS5A of Hepatitis C Virus Suggest Drug Resistance Mechanisms. Genes. 2018;9(7):343.
Crossref - Gaunt E, Wise HM, Zhang H, et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. Elife. 2016;5:e12735.
Crossref - Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID Validates Template Sampling Depth and Greatly Reduces the Error Rate of Next-Generation Sequencing of HIV-1 Genomic RNA Populations. Sundquist WI, editor. J Virol. 2015;89(16):8540-8555.
Crossref - Zhou Zh, Qiu Y, Ge X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim Dis. 2021;1:5.
Crossref - Rashed WM, Kandeil MAM, Mahmoud MO, Ezzat S. Hepatocellular Carcinoma (HCC) in Egypt: A comprehensive overview. J Egypt Natl Cancer Institute. 2020;32(1):35
Crossref - Mejer NF, Fahnoe U, Galli A, et al. Mutations Identified in the Hepatitis C Virus (HCV) Polymerase of Patients with Chronic HCV Treated with Ribavirin Cause Resistance and Affect Viral Replication Fidelity. Antimicrob Agents Chemother. 2020;64(12):e01417-20.
Crossref - Che Noh I, Avoi R, Nurul AA, Ahmad I, Bakar RA. Analysis of serum and gene expression profile of cytokines (IL-6, TNF-a and TGF-a1) in chronic hepatitis C virus infection. PeerJ. 2022;10:e13330.
Crossref - Zghair AN, Tektook NK, Mohsin AA. Expression of IL-1b, IL-12, IL-17a, TNF-a and Fox-P3 in Patients with Low, Medium and High-Hepatitis Viral Load. Ann Rom Soc Cell Biol. 2021;9239-9250.
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.