ISSN: 0973-7510
E-ISSN: 2581-690X
Fluoride contamination in water sources represents a persistent and grave global issue with severe health implications. Microorganisms are known to demonstrate tolerance to various pollutants, enabling their survival in diverse and adverse environments. In this study, we aimed to identify potential bacterial isolates capable of resisting and removing fluoride from contaminated water, and can be used for bioremediation of fluoride contaminated water The unique Coringa Wildlife Sanctuary, a mangrove forest, served as a collection site for soil, sludge and water samples due to its distinctive ability to host halotolerant and halophilic bacteria, setting it apart from other plant species in naturally saline conditions. This exceptional microbial community within mangroves established them as a valuable source of fluoride resistant bacteria. A total of 46 bacterial isolated from sludge, soil, and water samples within the Coringa Wildlife Sanctuary belonged to the families Bacillaceae, Vibrionaceae, and Enterobacteriaceae. Predominantly, sludge samples yielded the highest number of isolates (41%), followed by soil (33%) and water (26%). All isolates demonstrated varying degrees of fluoride resistance, with 57% tolerating concentrations up to 2000 mg/L. Further screening showed that 22% of isolates tolerated 10,000 mg/L fluoride, while none survived at 20,000 mg/L. Notably, strain MSO5 exhibited growth on 12,500 mg/L fluoride containing medium within 24 hours. Based on 16S rRNA molecular studies identified the fluoride-resistant isolate MSO5 as Bacillus paralicheniformis. This study marks the first report of strain MSO5 belonging to the Bacillus sp. exhibiting tolerance to 10% salt concentration, temperature 55 °C and resistance to fluoride upto 12,500 mg/L concentration. This research lays the foundation for isolating fluoride resistant bacteria capable of removing fluoride, providing valuable prospects for microbial remediation of contaminated water sources.
Fluoride Resistant Bacteria, Microbial Bioremediation, Bacillus, Coringa Mangrove Wildlife Sanctuary, Halophilic
The global challenge of providing clean and safe drinking water is exacerbated by the presence of numerous toxic pollutants, among them, fluoride. Fluorine, in the form of fluoride, ranks 13th in abundance on Earth, constituting approximately 0.09% of the planet’s surface. Industrial discharges, primarily as wastewater, are a major source of fluoride contamination in drinking water supplies.1
Fluoride contamination in groundwater typically originates from fluoride-rich minerals such as fluorite, biotite, topaz, fluorapatite, cryolite, hornblende, and muscovite, which release fluoride ions into water upon contact.2 Its concentration is influenced by environmental and geological factors, including aquifer conditions, rock weathering, water table depth, soil and rock acidity, porosity, and chemical interactions within aquifers.3,4
In drinking water, fluoride is beneficial below 0.5 mg/L but poses serious health risks above this threshold.5 High fluoride exposure (>2.0 mg/L) is linked to dental and skeletal fluorosis, crimping fluorosis, and osteosclerosis, affecting all age groups.6,7 Insufficient fluoride intake, on the other hand, can lead to dental caries, weak enamel, and bone fragility.8 Globally, around 260 million people suffer from fluorosis due to inadequate water treatment, with India, China, and Ethiopia being the most affected countries.9 Over 200 million individuals across 35 nations are at risk of fluorosis, highlighting the urgency for fluoride research.10
In southern India, fluoride-rich rocks are a major source of contamination, particularly in regions like Nalgonda, Andhra Pradesh, where granite fluoride levels exceed the global average of 810 mg/kg.11,12 In India, affected states include Andhra Pradesh, Bihar, Gujarat, Madhya Pradesh, Punjab, Rajasthan, Tamil Nadu, and Uttar Pradesh.13
Excess fluoride intake is the primary cause of dental and skeletal fluorosis. Levels above 2 mg/day can lead to mottled teeth and osteosclerosis, while prolonged exposure to 20 mg/day over 10-20 years may cause renal toxicity. Intake exceeding 6 mg/day significantly increases the risk of skeletal fluorosis.14 Acute fluorosis symptoms include nausea, vomiting, diarrhea, and cardiac arrhythmia, while skeletal fluorosis is characterized by joint stiffness and pain.13,15
The toxicity of fluoride (F”) differs significantly from that of these toxic elements, and F” is notoriously challenging to remove from water sources. Despite these challenges, several relatively cost-effective chemical and mechanical methods have been introduced for Fluoride removal, including electro-chemical processes, ion exchange methods and adsorption techniques.3 Investigators have reported certain advantages associated with F” removal methods that involve the accumulation or absorption of F” using various materials, such as activated carbon, alumina, and zeolites.16-18
Current study aimed to isolate fluoride resistant bacteria from natural sources particularly within unique ecological niches like mangrove habitats. This research paves the way for the development of innovative and sustainable solutions to address water quality challenges posed by fluoride contamination. Such microbial-based strategies hold possible bioremediation for enhancing environmental sustainability and safeguarding human health in regions struggling with fluoride pollution in their water supplies.19
The present study was carried out during 2023-2024 at Department of Biochemistry, Acharya Nagarjuna University, Nagarjunanagar, Guntur, Andhra Pradesh.
Area selection for possible isolation of fluoride resistant bacteria
Mangroves, as the primary productive marine ecosystems on our planet, offer a unique and distinct habitat that benefits to numerous species.20 These ecosystems are not only valuable in terms of their ecological significance but also serve as a source of untapped biological compounds that can be advantageous to both humans and plants.
Coringa wildlife Sanctuary, a mangrove forest was selected for collecting soil and water samples, because of its distinguishing characteristic of mangroves is their ability to harbor halotolerant and halophilic bacteria, unlike other plant species that grow in naturally saline conditions. This unique microbial community makes mangroves a valuable source of fluoride resistant bacteria (FRB).
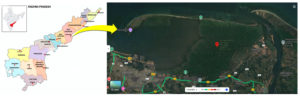
Samples were collected from three sources namely soil, sludge and water at different locations of mangrove forest located at Coringa Wildlife Sanctuary, East Godavari district within latitude of 16°49’53.0″N and longitude of 82°20’12.0″E (16.831389, 82.336667) (Figure 1).
Figure 1. Satellite view of Corangi Mangrove Wildlife Sanctuary, Kakinada.
Source: Google Maps at the coordinates 16°49’53.0″N (latitude) and 82°20’12.0″E (longitude) (16.831389, 82.336667). Accessed date 27 March 2024
Sample collected from soil
Soil samples were collected systematically from various locations within the Corangi Mangrove Wildlife Sanctuary, targeting the upper soil layer (5-10 cm depth) where microbial activity and bacterial populations are most concentrated. Debris was cleared from the soil surface before collection. Approximately 20 g of soil was sampled and stored in an icebox for transport to the laboratory.21,22
Sample collected from water
Water samples were aseptically collected from different locations within the Corangi Mangrove Wildlife Sanctuary using sterile 500 mL polythene bottles, submerged directly into the water’s surface. The samples were appropriately labeled and transported on ice for laboratory analysis. Aliquots of the water samples were used for the selective isolation of fluoride-resistant bacteria.21-23
Sample collected from Sludge
Sediment samples were collected from depths of 5 cm and 40 cm within the root zone of various plants. The samples were labeled and transported on ice to the laboratory. Aliquots were used for the selective isolation of fluoride-resistant bacteria.21-23
Isolation of Bacteria strain from soil, sludge and water
Glassware was cleaned and pre-treated with 10% (v/v) hydrochloric acid before use. Zobell Marine Agar plates (Zobell-Marine Broth 2216 Himedia at the rate 5.5% + Agar-agar at the rate 1.5%) were prepared by autoclaving at 121 °C and 15 psi for 15 minutes.
The standard serial dilution technique was employed for bacterial isolation from the samples. For soil, 1 g was mixed with 10 mL of sterile water and serially diluted (10-1 to 10-4). From the diluted samples, 100 µL was mixed with warm Zobell Marine Agar medium and poured into Petri plates. After 48 hours of incubation, mixed bacterial colonies formed a lawn. Individual colonies were picked using sterile toothpicks and streaked onto fresh nutrient agar plates to obtain pure cultures.24
Maintenance of bacterial cultures
The cultures were stored for long-term preservation using glycerol stock at refrigerated conditions (-4 °C). Purified isolates were maintained for extended periods at -20 °C in nutrient broth with 20% (w/v) glycerol. For short-term storage and further characterization, the cultures were stored on nutrient agar plates at 4 °C.23
Screening of bacteria for fluoride resistance
Screening of fluoride resistance bacteria was done according to the procedure outlined by Mukherjee, et al.25 Nutrient agar media with different concentrations of 25, 100, 500, 1,000, 2,000, 5,000, 10,000, 12,000 and 13,000 mg/L was prepared using NaF (sodium fluoride). The bacterial cultures were primarily streaked on low concentration of NaF and subsequently incubated at a temperature of 32 °C for a period of 24 hours. Following incubation, any microorganisms that exhibited resistance to fluoride were selected and streaked on high concentration of NaF containing medium.
Phenotypic characters of the bacterial isolates
Phenotypic characteristics including colony morphology, cell morphology, motility, endospore formation, and Gram staining reactions were documented. Biochemical assays, such as KOH, Catalase, Oxidase, and Amino Acid Decarboxylase tests, were conducted following the methodology outlined by Aneja.26 Furthermore, carbohydrate fermentation tests and IMViC assays, as described by Seeley and Vandemark,27 were utilized to partially identify the bacterial cultures.
Property studies of bacterial isolates
Salt tolerance
All isolated organisms underwent salt tolerance testing at varying concentrations of NaCl, including 0%, 6%, 8%, and 10%. Each organism was streaked onto Nutrient agar plates containing different salt concentrations and subsequently incubated at 32 °C. Growth observations were recorded at both 24-hour and 48-hour intervals.
Temperature tolerance
The isolated organisms were streaked onto 3% NaCl Nutrient agar plates and then subjected to incubation at different temperatures ranging from 40 °C, 45 °C, 50 °C, and 55 °C. Subsequent observations of growth were recorded at both 24-hour and 48-hour intervals.
Gelatin hydrolysis28
The gelatinase enzyme activity, indicative of gelatin hydrolysis, was assessed through gelatin liquefaction. The test cultures were inoculated into nutrient gelatin using a spot method and subsequently incubated at 32 °C for 48 hours. Following incubation, the tubes were observed for any signs of gelatin liquefaction by flooding with Ammonium suphate, Clear zone around the colonies indicating the presence of gelatinase activity.
Starch hydrolysis27
The starch hydrolysis test was conducted to assess the activity of amylase. Bacterial isolates were streaked onto nutrient agar plates supplemented with 2% insoluble starch, followed by incubation at room temperature. After incubation, the plates were flooded with iodine solution to visualize starch hydrolysis. The presence of clear zones surrounding the colonies indicated a positive reaction, indicative of starch hydrolysis.
Bacterial Identification by using 16S rRNA molecular method
DNA was extracted from the bacterial culture and assessed for quality on a 1.0% agarose gel, revealing a single band of high-molecular-weight DNA. Subsequently, the 16S rRNA gene fragment was amplified using 16S rRNA-F and 16S rRNA-R primers, yielding a distinct PCR amplicon band of 1500 bp when visualized on agarose gel. The PCR product was then purified to eliminate impurities before undergoing forward and reverse DNA sequencing reactions with the same primers. A consensus sequence of the 16S rRNA gene was generated by aligning the forward and reverse sequence data using aligner software. This sequence was subjected to BLAST analysis against the ‘nr’ database of the NCBI GenBank, and the top ten sequences with the highest identity scores were selected. These sequences were further aligned using the Clustal W multiple alignment software program. Finally, a distance matrix and phylogenetic tree were constructed using MEGA 10 based on the aligned sequences.
Isolation of bacterial culture from soil, sludge and water at different locations of mangrove forest
Overall 46 bacterial strains were isolated from three sources of mangrove forest distributed at Coringa Wildlife Sanctuary, Kakinada. Pure bacterial isolates were named concurrence to the area followed by source and finally by serial number as showed in Table 1.
Table (1):
Coding of bacterial strains isolated from different natural sources
| Area | Source | Bacterial codes |
|---|---|---|
| Coringa Mangrove Wildlife Sanctuary | Soil | MSO1, MSO2, MSO3, MSO4, MSO5, MSO6, MSO7, MSO8, MSO9, MSO10, MSO11, MSO12, MSO13, MSL14, MSL15 |
| Sludge | MSL1, MSL2, MSL3, MSL4, MSL5, MSL6, MSL7, MSL8, MSL9, MSL10, MSL11, MSL12, MSL13, MSL14, MSL15, MSL16, MSL17, MSL18, MSL19 | |
| Water | MW1, MW2, MW3, MW4, MW5, MW6, MW7, MW8, MW9, MW10, MW11, MW12 |
Note: M-Mangrove; SO-Soil; SL-Sludge; W-Water
Overall 46 bacterial strains were isolated from three sources of mangrove forest distributed at Coringa Wildlife sanctuary, Kakinada. Among them highest number of bacterial cultures were obtained from sludge (41%) followed by soil (33%) and water (26%). Percentage distribution and occurrence of bacterial cultures among difference source were showed in Table 2.
Table (2):
Percentage distribution and occurrence of endophytic bacteria between selected mangrove plant sp.
Source |
Bacterial isolates |
Percentage distribution |
|---|---|---|
Soil |
15 |
33 % |
Sludge |
19 |
41 % |
Water |
12 |
26 % |
Total isolates |
46 |
Screening of Fluoride tolerant bacteria (FTB)
Total of 46 bacterial cultures obtained are screened to fluoride resistant as per the method described in Materials and Methods.
All the isolates showed varying degree of resistance to fluoride. Preliminary the bacterial isolates tested for fluoride tolerance to low concentration of fluoride. Among them 72% showed positive growth, while 28% showed negative growth on 25 mg/L concentration. Furthermore, 61% showed fluoride tolerance to 100 mg/L concentration, while 59% of bacterial isolates showed growth on 500 mg/L and 57% of bacterial isolates showed positive growth on 1000 to 2000 mg/L concentrations (Table 3).
Table (3):
Fluoride screening of microorganisms at low concentrations
| No. | Source | Organisms | Type of organisms | 25 mg/L | 100 mg/L | 500 mg/L | 1000 mg/L | 2000 mg/L |
|---|---|---|---|---|---|---|---|---|
| 1. | Soil (15) | MSO1 | Vibrio | + | − | − | − | − |
| 2. | MSO2 | Bacillus | − | − | − | − | − | |
| 3. | MSO3 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 4. | MSO4 | Vibrio | − | − | − | − | − | |
| 5. | MSO5 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 6. | MSO6 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 7. | MSO7 | Bacillus | + | + | − | − | − | |
| 8. | MSO8 | Vibrio | − | − | − | − | − | |
| 9. | MSO9 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 10. | MSO10 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 11. | MSO11 | Vibrio | − | − | − | − | − | |
| 12. | MSO12 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 13. | MSO13 | Pseudomonas | + | − | − | − | − | |
| 14. | MSO14 | Bacillus | + | − | − | − | − | |
| 15. | MSO15 | Vibrio | ++ | ++ | ++ | ++ | ++ | |
| 16. | Sludge (19) | MSL1 | Vibrio | ++ | ++ | ++ | ++ | ++ |
| 17. | MSL2 | Vibrio | ++ | ++ | ++ | ++ | ++ | |
| 18. | MSL3 | Bacillus | − | − | − | − | − | |
| 19. | MSL4 | Bacillus | − | − | − | − | − | |
| 20. | MSL5 | Vibrio | ++ | ++ | ++ | ++ | ++ | |
| 21. | MSL6 | Virbio | − | − | − | − | − | |
| 22. | MSL7 | Aeromonas | + | − | − | − | − | |
| 23. | MSL8 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 24. | MSL9 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 25. | MSL10 | Enterobacter | − | − | − | − | − | |
| 26. | MSL11 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 27. | MSL12 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 28. | MSL13 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 29. | MSL14 | Bacillus | − | − | − | − | − | |
| 30. | MSL15 | Enterobacter | − | − | − | − | − | |
| 31. | MSL16 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 32. | MSL17 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 33. | MSL18 | Vibrio | + | − | − | − | − | |
| 34. | MSL19 | Bacillus | ++ | ++ | ++ | ++ | ++ | |
| 35. | Water (12) | MW1 | Bacillus | ++ | ++ | ++ | ++ | ++ |
| 36. | MW2 | Aeromonas | + | + | + | − | − | |
| 37. | MW3 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 38. | MW4 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 39. | MW5 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 40. | MW6 | Enterobacter | − | − | − | − | − | |
| 41. | MW7 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 42. | MW8 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 43. | MW9 | Vibrio | − | − | − | − | − | |
| 44. | MW10 | Bacillus | − | − | − | − | − | |
| 45. | MW11 | Bacillus | ++ | ++ | ++ | ++ | + | |
| 46. | MW12 | Bacillus | ++ | ++ | ++ | ++ | + |
Note: “++” represents high growth; “+” represents moderate growth, and “−” represents no growth. The results were obtained are the mean values of three replicates.
Following the initial screening, a secondary screening process was conducted to assess fluoride tolerance at higher concentrations. Of the 26 bacterial cultures displaying fluoride tolerance up to 2000 mg/L, a total of 21 isolates (81%) exhibited positive growth in the presence of 5000 mg/L fluoride. Furthermore, 10 isolates (38%) demonstrated remarkable tolerance, surviving concentrations of up to 10,000 mg/L. Among them strain MSO5 showed fluoride resistance, withstanding concentrations of upto 12,500 mg/L, making it a promising candidate for further investigation. Notably, none of the bacterial isolates were viable at the exceptionally high fluoride concentration of 20,000 mg/L (Table 4).
Table (4):
Fluoride screening of microorganisms at high concentrations
| No. | Source | Bacterial isolates | Type of organisms | 5000 mg/L | 10,000 mg/L | 12,500 mg/L | 20,000 mg/L |
|---|---|---|---|---|---|---|---|
| 1. | Soil (07) | MSO3 | Bacillus | ++ | − | − | − |
| 2. | MSO5 | Bacillus | ++ | + (24 hrs) | + (24 hrs) | − | |
| 3. | MSO6 | Bacillus | ++ | + (48 hrs) | − | − | |
| 4. | MSO9 | Bacillus | ++ | + (48 hrs) | − | − | |
| 5. | MSO10 | Bacillus | ++ | − | − | − | |
| 6. | MSO12 | Bacillus | ++ | − | − | − | |
| 7. | MSO15 | Vibrio | ++ | + | − | − | |
| 8. | Sludge (11) | MSL1 | Vibrio | ++ | + | − | − |
| 9. | MSL2 | Vibrio | ++ | + | − | − | |
| 10. | MSL5 | Vibrio | ++ | + | − | − | |
| 11. | MSL8 | Bacillus | + | − | − | − | |
| 12. | MSL9 | Bacillus | + | − | − | − | |
| 13. | MSL11 | Bacillus | + | − | − | − | |
| 14. | MSL12 | Bacillus | − | − | − | − | |
| 15. | MSL13 | Bacillus | + | + (48 hrs) | − | − | |
| 16. | MSL16 | Bacillus | + | + (48 hrs) | − | − | |
| 17. | MSL17 | Bacillus | − | − | − | − | |
| 18. | MSL19 | Bacillus | − | − | − | − | |
| 19. | Water (08) | MW1 | Bacillus | + | − | − | − |
| 20. | MW3 | Bacillus | + (just survive) | − | − | − | |
| 21. | MW4 | Bacillus | + (just survive) | − | − | − | |
| 22. | MW5 | Bacillus | ++ | + (48 hrs) | − | − | |
| 23. | MW7 | Bacillus | + (just survive) | − | − | − | |
| 24. | MW8 | Bacillus | + (just survive) | − | − | − | |
| 25. | MW11 | Bacillus | + (just survive) | − | − | − | |
| 26. | MW12 | Bacillus | − | − | − | − |
Note: “++” represents high growth; “+” represents moderate growth, and “−” represents no growth. The results were obtained are the mean values of three replicates.
Salt and temperature tolerance
Bacterial strain MSO5 showed fluoride tolerance upto 10,000 mg/L and exhibited growth within 24 hours of incubation. This strain also tolerant salt upto 10% NaCl concentration in media. This strain was also tested for temperature tolerance and the results showed that strain MSO5 was tolerant and exhibit growth upto 55 °C of temperature.
Enzyme production
All isolates were screened for Amylase and Protease enzyme production. Among them, bacterial strain MSO5 showed highest amylolactic index of 3.1 cm and proteolytic index of 3.5 cm.
Phenotypic characters of potential bacteria strain
The bacterial strain MSO5 isolated from the sample exhibits several characteristic features indicative of its identity and potential behavior. Microscopic observation showed the presence of rod-shaped bacteria occurring in pairs. Gram staining demonstrated a positive result, suggesting the strain’s cell wall composition. Furthermore, the presence of endospores was observed, indicating the strain’s ability to form dormant structures for survival in unfavorable conditions. The cells were oval in shape, positioned at the center of the cell. However, motility was absent, suggesting a lack of flagella for movement. Biochemical tests showed positive results for catalase and oxidase activities, indicative of metabolic capabilities. Notably, MSO5 displayed robust growth at elevated temperatures, thriving excellently at both 42 °C and 55 °C. Starch and gelatin hydrolysis tests yielded positive results, suggesting the strain’s ability to utilize these substrates. Moreover, MSO5 exhibited varying degrees of tolerance to NaCl, with notable growth observed at concentrations up to 6%. Strong hemolysis was observed, indicating potential pathogenicity. On Bacillus selective media, the strain displayed a characteristic green coloration. Additional biochemical tests revealed positive results for urease and arginine decarboxylase, among others, further supporting the identification of the strain as a member of the Bacillus genus. Overall, the combined results from microscopic, biochemical, and growth characteristic tests preliminarily identify the isolated strain as Bacillus sp., emphasizing its potential for further study and application in various fields, including bioremediation.
Microbial Identification using 16S rRNA gene based molecular method
Overall 10 bacterial isolates was exhibited fluoride resistance upto 10,000 mg/L. Among them MSO5 was selected for molecular studies because of its ability to absorb fluoride, able to tolerate high concentration of salt and temperature when compared to other test isolates fluoride resistance strains.
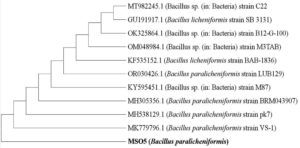
The DNA extracted from MSO5 cultures showed a single high molecular weight DNA band on agarose gel electrophoresis. Amplification of the 16S rRNA gene yielded a 1500 bp PCR amplicon. BLAST analysis revealed a 100% similarity between MSO5 (Nearest Accession no. MK779796.1) and Bacillus paralicheniformis. Phylogenetic analysis confirmed the clustering of MSO5 with Bacillus paralicheniformis, forming a distinct group separate from other closely related species (Table 5 and Figure 2).
Table (5):
Significant alignments of Sequence produced
Particualrs |
Maximum Score |
Total Score |
Query Cover |
E Value |
Percent Identity |
Nearest Accessions |
|---|---|---|---|---|---|---|
Bacillus paralicheniformis strain VS-1 |
2747 |
2747 |
100% |
0.0 |
100.00% |
MK779796.1 |
Bacillus paralicheniformis strain pk7 |
2747 |
2747 |
100% |
0.0 |
100.00% |
MH538129.1 |
Bacillus paralicheniformis strain BRM043907 |
2745 |
2745 |
99% |
0.0 |
100.00% |
MH305356.1 |
Bacillus sp. (in: Bacteria) strain M87 |
2745 |
2745 |
99% |
0.0 |
100.00% |
KY595451.1 |
Bacillus paralicheniformis strain LUB129 |
2745 |
2745 |
99% |
0.0 |
100.00% |
OR030426.1 |
Bacillus licheniformis strain BAB-1836 |
2745 |
2745 |
99% |
0.0 |
100.00% |
KF535152.1 |
Bacillus sp. (in: Bacteria) strain M3TAB |
2743 |
2743 |
99% |
0.0 |
100.00% |
OM048984.1 |
Bacillus sp. (in: Bacteria) strain B12-G-100 |
2741 |
2741 |
99% |
0.0 |
100.00% |
OK325864.1 |
Bacillus sp. (in: Bacteria) strain C22 |
2741 |
2741 |
99% |
0.0 |
100.00% |
MT982245.1 |
Bacillus licheniformis strain SB 3131 |
2741 |
2741 |
99% |
0.0 |
99.93% |
GU191917.1 |
The current study underscores the potential of bioremediation technologies in addressing fluoride contamination issues effectively. While, various approaches such as, nanofiltration,28 membrane separation technique,29 electro-coagulation method,30 reverse osmosis,31 adsorption,32 ion exchange process33 Nalgonda method, particle trade34 etc., have been utilized for defluoridation of potable water.35 However, these methods have drawbacks, including high costs, energy consumption, and the generation of secondary contaminants post-treatment. Additionally, they may not efficiently eliminate all contaminants present in water and wastewater.36 In contrast, microorganisms offer distinct advantages, including ease of operation and lower sludge production, making them a promising alternative for water treatment and wastewater remediation.
The strong fluoride resistance displayed by Bacillus paralicheniformis strain MSO5 suggests its potential for treating fluoride-contaminated water. While this study focused mainly on identifying fluoride resistance, it also facilitates the feasibility of developing a bioremediation system. Methods such as immobilization of bacterial cultures to prevent washout or implementing sequence batch reactors could enhance the longevity and efficacy of the bacteria. Further experiments are underway to assess the sustainability and practicality of these bioremediation approaches. Prior research on bioremediation has consistently highlighted the versatile and effective application of Bacillus species in the cleanup of diverse pollutants.37,38
The present study represents the first report of Bacillus paralicheniformis strain MSO5 demonstrating fluoride tolerance of up to 12,500 mg/L. Additionally, it is the first documented instance of the Bacillus genus exhibiting fluoride resistance, contrasting with previously reported genera such as Pseudomonas, Enterobacter, and Aeromonas (Table 6).
Table (6):
Comparative study showing Bacillus paralichiniformis strain MSO5 with other bacterial isolates
| Bacteria Name | Fluoride resistant In mg/L (PPM) | Equivalent to In mM | Reference |
|---|---|---|---|
| Bacillus paralichiniformis strain MSO5 | 12,500 mg/L | 240 mM | Present work |
| Pseudomonaas aerugenosa | 12,957 mg/L | 300 mM | Chellaiah et al.39 |
| Enterobacter cloacae | 8398-12597 mg/L | 200-300 mM | |
| Aeromonas, E. coli and Enterobacter species | 8398-12597 mg/L | 200-300 mM | |
| Aeromonas sp. | 9200 mg/L | 221 mM | Sharma et al.40 |
| Pseudomonas sp. | 8398 mg/L | 200 mM | Sree et al.41 |
| Pseudomonas aeruginosa | 8398 mg/L | 200 mM | Edward, et al.39 |
| Brevibacterium sp. | 7200 mg/L | 173 mM | Sharma et al.40 |
| Paenibacillus sp. | 5200 mg/L | 125 mM | Sharma et al.40 |
| Micrococcus luteus, Aeromonas hydrophyla, Micrococcus varians, Pseudomonas aerugenosa | 4199 mg/L | 100 mM | Chouhan, et al.42 |
| Bacillus australimaris NH71_1 | 4000 mg/L | 96 mM | Thirumala, et. al.43 |
| Bacillus cereus FT1 Bacillus marisflavi FT2 |
3000 mg/L | 71.4 mM | Banerjee, et. al.44 |
| Bacillus flexus | 500-2500 mg/L | 12- 60 mM | Thesai et al.45 |
| Streptococcus mutants UA159FR | 1,000 mg/L | 52.6 mM | Liao, et al.46 |
| Bacillus megaterium | 1500 mg/L | 36 mM | Pal, et al.47 |
| Bacillus flexus | 1500 mg/L | 35.7 mM | Pal, et al.48 |
| Acinetobacter sp. RH5 | 250 mg/L | 6 mM | Mukherjee, et al.25 |
*Note: Formula for conversion of Fluoride concentration (Molecular weight of NaF is 41.98 g/mol) from mg/L to mM is 1 milli molar = 41.99 mg/L (ppm)
The salinity of the growth medium plays a pivotal role in the viability and metabolic activities of bacteria. Variations in salt concentrations between the external environment and the bacterial cytoplasm create osmotic pressure on the bacterial cell wall, and excessive pressure can lead to cell membrane disruption. Furthermore, alterations in salinity levels can impact the activity of various enzymes within bacterial cells, as previously noted.16
It is important to recognize that the salinity of the surrounding environment may have a significant influence on the bioremediation capacity of bacteria. In our study, we identified a bacterial isolate capable of withstanding a broad spectrum of salt concentrations in the growth medium. This isolate exhibited growth even in the presence of salt concentrations spanning from 3% to 10% (w/v). Previous studies have also reported the resilience of certain Bacillus species to high salt concentrations, where such conditions had minimal detrimental effects on their growth. These findings underscore the adaptability of specific bacterial strains to varying salinity levels, which can have important implications for their utility in bioremediation processes and various applications in saline environments.
In our present study, we have characterized one bacterial strains, namely Bacillus licheniformis strain MSO5 which was isolated from soil sample at Coringa wildlife mangrove ecosystem. This bacterial strains demonstrated a remarkable tolerance to high concentrations of F” in the form of NaF. Bacillus licheniformis is a Gram-positive bacterium that resides in soil, forms spores, and can thrive in both aerobic and anaerobic conditions. Bacteria employ various mechanisms to cope with toxic substances, and in the case of F”, they possess a unique channel protein referred to as putative F-transporters, which help mitigate the toxic effects of F” on bacterial cells.49 These transporters are, in fact, riboswitches-special metabolite-binding RNA structures that are activated in the presence of high F” concentrations. Additionally, it has been observed that these channel transporters exhibit a strong affinity for fluoride ions while excluding other negatively charged ions, such as chloride.50 This study not only expands our understanding of bacterial adaptation to challenging environments but also underscores the potential of these bacterial strains in addressing F contamination, offering promising solutions for the treatment of fluoride-polluted waters.
The isolation of Bacillus paralicheniformis strain MSO5 from the Coringa Mangrove Wildlife Sanctuary represents a significant finding, as it exhibits remarkable resistance to fluoride concentrations of up to 12,500 mg/L, along with its ability to tolerate 10% salt concentration and grow at a temperature of 55 °C. This study marks species level identification of a member of the Bacillus family, namely Bacillus paralicheniformis strain MSO5, demonstrating such high tolerance to fluoride. These findings underscore the potential of this strain as a valuable candidate for the development of microbial remediation strategies to combat fluoride contamination in various water sources.
ACKNOWLEDGMENTS
The authors express their sincere gratitude to the Department of Biochemistry, Acharya Nagarjuna University, Guntur, for providing the necessary facilities to carry out this research work. The authors also extend their appreciation to the staff of For U International Private Limited, Research & Development Laboratories, Visakhapatnam, for their technical expertise and support in sample collection.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Shanker AS, Srinivasulu D, Pindi PK. A study on bioremediation of fluoride-contaminated water via a novel bacterium Acinetobacter sp.(GU566361) isolated from potable water. Results Chem. 2020;2:100070.
Crossref - Rizzu M, Tanda A, Cappai C, Roggero PP, Seddaiu G. Impacts of soil and water fluoride contamination on the safety and productivity of food and feed crops: A systematic review. Sci Total Environ. 2021;787:147650.
Crossref - Raj D, Shaji E. Fluoride contamination in groundwater resources of Alleppey, southern India. Geosci Front. 2017;8(1):117-124.
Crossref - Raju NJ, Dey S, Gossel W, Wycisk P. Fluoride hazard and assessment of groundwater quality in the semi-arid Upper Panda River basin, Sonbhadra district, Uttar Pradesh, India. Hydrological Sciences Journal. 2012;57(7):1433-1452.
Crossref - Biswas G, Thakurta SG, Chakrabarty J, Adhikari K, Dutta S. Evaluation of fluoride bioremediation and production of biomolecules by living cyanobacteria under fluoride stress condition. Ecotoxicol Environ Safety. 2018;148:26-36.
Crossref - Affonso LN, Marques Jr JL, Lima VVC, et al. Removal of fluoride from fertilizer industry effluent using carbon nanotubes stabilized in chitosan sponge. J Hazard Mater. 2020;388:122042.
Crossref - Shaji E, Sarath KV, Santosh M, Krishnaprasad PK, Arya BK, Babu MS. Fluoride contamination in groundwater: A global review of the status, processes, challenges, and remedial measures. Geoscience Frontiers. 2024;15(2):101734.
Crossref - Bhatnagar A, Kumar E, Sillanpaa M. Fluoride removal from water by adsorption-a review. Chem Eng J. 2011;171(3):811-840.
Crossref - World Health Organization. Guidelines for drinking-water quality: incorporating the first and second addenda. World Health Organization. 2022.
- Kabir H, Gupta AK, Tripathy S. Fluoride and human health: Systematic appraisal of sources, exposures, metabolism, and toxicity. Crit Rev Environ Sci Technol. 2020;50(11):1116-1193.
Crossref - Brindha K, Elango LX. Fluoride in groundwater: causes, implications and mitigation measures. In: Monroy SD Fluoride Properties, Applications and Environmental Management. 2011;1:111-136.
- Tikki MA. Fluoride removal from water-a review. Int J Sci Eng Res. 2014;5(1):515-519.
- Singh A, Gothalwal R. A reappraisal on biodegradation of fluoride compounds: role of microbes. Water Environ J. 2018;32(3):481-487.
Crossref - Fewtrell L, Smith S, Kay D, Bartram J. An attempt to estimate the global burden of disease due to fluoride in drinking water. J Water Health. 2006;4(4):533-542.
Crossref - Gazzano E, Bergandi L, Riganti C, et al. Fluoride effects: the two faces of janus. Curr Med Chem. 2010;17(22):2431-2441.
Crossref - Diaz-Nava C, Olguin MT, Solache-Rios M. Water defluoridation by Mexican heulandite-clinoptilolite. Separ Sci Technol. 2002;37(13):3109-3128.
Crossref - Jimenez-Nunez MML, Solache-Rios M, Olguin MT. Fluoride removal from aqueous solutions by magnesium, nickel, and cobalt calcined hydrotalcite-like compounds. Separ Sci Technol. 2010;45(6):786-793.
Crossref - Ghorai S, Pant KK. Equilibrium, kinetics and breakthrough studies for adsorption of fluoride on activated alumina. Separ Sci Technol. 2005;42(3):265-271.
Crossref - Chouhan S, Tuteja U, Flora SJS. Isolation, identification and characterization of fluoride resistant bacteria: possible role in bioremediation. ApplBiochem Microbiol. 2012;48:43-50.
Crossref - Carugati L, Gatto B, Rastelli E, et al. Impact of mangrove forests degradation on biodiversity and ecosystem functioning. Sci Rep. 2018;8(1):13298.
Crossref - Mulamattathil SG, Bezuidenhout C, Mbewe M, Ateba CN. Isolation of environmental bacteria from surface and drinking water in Mafikeng, South Africa, and characterization using their antibiotic resistance profiles. J Pathog. 2014;2014(1):371208.
Crossref - Pushkar B, Sevak P, Sounderajan S. Assessment of the bioremediation efficacy of the mercury resistant bacterium isolated from the Mithi River. Water Supply. 2019;19(1):191-199.
Crossref - Divakar G, Sameer RS, Bapuji M. Screening of multi-metal tolerant halophilic bacteria for heavy metal remediation. Int J Curr Microbiol App Sci. 2018;7(10):2062-2076.
Crossref - Prashanthi R, Shreevatsa GK, Krupalini S, Manoj L. Isolation, characterization, and molecular identification of soil bacteria showing antibacterial activity against human pathogenic bacteria. J Genet Eng Biotechnol. 2021;19(1):120.
Crossref - Mukherjee S, Sahu P, Halder G. Microbial remediation of fluoride-contaminated water via a novel bacterium Providencia vermicola (KX926492). J Environ Manag. 2017;204(1):413-423.
Crossref - Aneja KR. Experiments in Microbiology, Plant Pathology and Biotechnology. 4th Edition. New Delhi. 2006:245-275.
- Seeley HW, Vandemark PJ, Lee JJ. Microbes in action. A Laboratory Manual of Microbiology. Freeman and Company, San Francisco, USA. 1981:388.
- Jadhav SV, Marathe KV, Rathod VK. A pilot scale concurrent removal of fluoride, arsenic, sulfate and nitrate by using nanofiltration: Competing ion interaction and modelling approach. J Water Process Eng. 2016;13:153-167.
Crossref - Ndiaye PI, Moulin P, Dominguez L, Millet JC, Charbit F. Removal of fluoride from electronic industrial effluent by RO membrane separation. Desalination. 2005;173(1):25-32.
Crossref - Sandoval MA, Fuentes R, Nava JL, Rodriguez I. Fluoride removal from drinking water by electrocoagulation in a continuous filter press reactor coupled to a flocculator and clarifier. Sep Purif Technol. 2014;134:163-170.
Crossref - Colla V, Branca TA, Rosito F, Lucca C, Vivas BP, Delmiro VM. Sustainable reverse osmosis application for wastewater treatment in the steel industry. J Clean Prod. 2016;130:103-115.
Crossref - Ali I. Water treatment by adsorption columns: evaluation at ground level. Separation & Purification Reviews. 2014;43(3):175-205.
Crossref - Markovski J, Garcia J, Hristovski KD, Westerhoff P. Nano-enabling of strong-base ion-exchange media via a room-temperature aluminum (hydr) oxide synthesis method to simultaneously remove nitrate and fluoride. Sci Total Environ. 2017;599-600:1848-1855.
Crossref - Tirkey P, Bhattacharya T, Chakraborty S. Optimization of fluoride removal from aqueous solution using Jamun (Syzygium cumini) leaf ash. Process Saf Environ Prot. 2018;115:125-138.
Crossref - Gwala P, Andey S, Mhaisalkar V, Labhasetwar P, Pimpalkar S, Kshirsagar C. Lab scale study on electrocoagulation defluoridation process optimization along with aluminium leaching in the process and comparison with full scale plant operation. Water Sci Technol. 2011;63(12):2788-2795.
Crossref - Gentili FG, Fick J. Algal cultivation in urban wastewater: an efficient way to reduce pharmaceutical pollutants. J Appl Phycol. 2017;29(1):255-262.
Crossref - Guo H, Luo S, Chen L, et al. Bioremediation of heavy metals by growing hyperaccumulator endophytic bacterium Bacillus sp. L14. Bioresour Technol. 2010;101(22):8599-8605.
Crossref - Pandey S, Ghosh PK, Ghosh S, De TK, Maiti TK. Role of heavy metal resistant Ochrobactrum sp. and Bacillus spp. strains in bioremediation of a rice cultivar and their PGPR like activities. J Microbiol. 2013;51:11-17.
Crossref - Chellaiah ER, Ravi P, Uthandakalaipandian R. High fluoride resistance and virulence profile of environmental Pseudomonas isolated from water sources. Folia Microbiol. 2021;66(4):569-578.
Crossref - Sharma S, Upadhyay D, Singh B, Shrivastava D, Kulshreshtha NM. Defluoridation of water using autochthonous bacterial isolates. Environ Monit Assess. 2019;191(12):781.
Crossref - Sree KK, Raja CE, Ramesh U. Isolation and characterization of fluoride resistant bacteria from groundwaters in Dindigul, Tamilnadu, India. Environ Res Technol. 2018;1(2):69-74.
- Chouhan S, Tuteja U, Flora SJS. Isolation, identification and characterization of fluoride resistant bacteria: possible role in bioremediation. Appl Biochem Microbiol. 2012;48:43-50.
Crossref - Thirumala M, Krishna ES, Priya PS, Reddy SV. Characterization of a novel Fluoride resistant bacterial isolate and its capability of Fluoride bioremediation. AIMS Microbiol. 2022;8(4):470-483.
Crossref - Banerjee G, Sengupta A, Roy T, Banerjee PP, Chattopadhyay A, Ray AK. Isolation and characterization of fluoride resistant bacterial strains from fluoride endemic areas of west Bengal, India: assessment of their fluoride absorption efficiency. Fluoride. 2016;49(4):429.
- Thesai AS, Rajakumar S, Ayyasamy PM. Removal of fluoride in aqueous medium under the optimum conditions through intracellular accumulation in Bacillus flexus (PN4). Environmental technology;2018:41:1185-1198.
- Liao Y, Brandt BW, Li J, Crielaard W, Van Loveren C, Deng DM. Fluoride resistance in Streptococcus mutans: a mini review. J Oral Microbiol. 2017;9(1):1344509.
Crossref - Pal KC, Mukhopadhyay P, Chatterjee S, Mondal NK. A study on fluoride bioremediation via a novel bacterium Bacillus megaterium (JF273850) isolated from agricultural soil. J Earth Syst Sci. 2022;131(3):183.
Crossref - Pal KC, Mondal NK, Chatterjee S, Ghosh TS, Datta JK. Characterization of fluoride-tolerant halophilic Bacillus flexus NM25 (HQ875778) isolated from fluoride-affected soil in Birbhum District, West Bengal, India. Environ Monit Assess. 2014;186:699-709.
Crossref - Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335(6065):233-235.
Crossref - Praveen Kumar V, Hari Priya VR. Molecular characterization of fluorine degrading bacteria from soil samples for its industrial exploitation. Int J Adv Life Sci. 2013;6(4):351-355.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.