The only species of Entamoeba considered pathogenic untill now was Entamoeba histolytica. However, recent studies have highlighted the pathogenic potential and increasing gastrointestinal infections associated with E. moshkovskii. This study describes the molecular detection of E. moshkovskii in a symptomatic patient, indicating its possible prevalence across India. As this species appears to be an emerging pathogen in the Indian subcontinent, further epidemiological research involving diverse patient cohorts is necessary to assess its disease incidence. This will aid in understanding the epidemiology and clinical significance of E. moshkovskii, thereby improving diagnosis and management strategies for amoebiasis.
Entamoeba moshkovskii, Molecular Detection, Emerging Pathogen, North India
The World Health Organization (WHO) has ranked amoebiasis as the third major reason of mortality among parasitic ailments, accounting for over 100,000 deaths per year, especially in developing countries. The disease is prevalent in areas with poor sanitation and the cysts of Entamoeba histolytica, are easily acquired through the ingestion of contaminated food and water.1 Clinical manifestations of amoebiasis can vary from asymptomatic infections to severe signs, including the development of extraintestinal abscesses.2 The Entamoeba genus includes several other species that inhabit the intestinal lumen. Previous studies have shown that patients with gastrointestinal symptoms may be infected with E. moshkovskii and E. dispar, but establishing their pathogenic potential requires further conclusive evidence.3 The challenge lies in the fact that these three species are morphologically similar, making them indistinguishable by microscopy alone. E. moshkovskii was first described in 1941 by Tshalai and was initially isolated from sewage water in Moscow.4,5 In India, it was first reported in patients with dysentery/diarrhea and abdominal discomfort from Pondicherry, South India.3 The species has further been detected in HIV patients from Assam and in diarrheal patients from Kolkata.6-8 A literature review indicated that the species has not been previously reported in North India and, therefore, is not routinely screened.9 Given this context, stool samples submitted for routine microscopy in the Department of Medical Parasitology were systematically screened for Entamoeba infection as part of a study aimed at detecting the presence of E. moshkovskii. Here, we report the detection of E. moshkovskii via molecular techniques from the North Indian region.
The study was approved by the Institutional Ethics Committee, Post Graduate Institute of Education and Research (PGIMER) Chandigarh, India, vide letter number INT/IEC/2022/SPL-766 dated 16.04.2022. Written informed consent was obtained from the participants before enrolling in the study.
A 57-year-old male patient, a native of Uttarakhand, presented to the Gastroenterology Outpatient Department (OPD) with complaints of epigastric and abdominal pain and altered bowel habits, persisting for the past 4 months. He also experienced postprandial fullness and reported a history of borborygmi. The patient was prescribed Mebeverine HCL & Chlordiazepoxide tablets along with Pantoprazole (40 mg) for two weeks on presenting in the OPD. The routine blood investigations, including a complete blood count, showed results within the normal range, except for a slightly elevated eosinophil percentage of 7.8%. An ultrasound examination of the entire abdomen revealed no abnormalities. The stool microscopy was negative for any other intestinal parasite and there were no observed pus cells or red blood cells. Given the low sensitivity of microscopy, negative samples were further processed using molecular techniques. The patient continued to experience occasional crampy pain in the abdomen and postprandial fullness. Bacterial stool culture and PCR for E. histolytica were also negative. He was further prescribed Rabeprazole for two weeks followed by Chlordiazepoxide and Clidinium Bromide tablets for a month after which he returned to his hometown and was lost to follow-up before the confirmation of infection via sequencing.
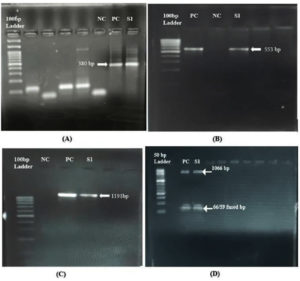
The Mini Paracep SF faecal concentrator was used to process the stool sample according to the manufacturer’s instructions. The pellet obtained was used for the preparation of a simple wet mount and an iodine-stained mount which was examined under the microscope.10 Lyophilized DNA of the E. moshkovskii Laredo strain was used as a positive control, generously provided by Professor Shinjiro Hamano and Professor Shin-Ichi Inoue, Department of Parasitology, Nagasaki University, Japan. DNA was extracted from the stool sample using QiAamp® DNA stool Mini kit (Qiagen, Hilden, Germany) with slight modifications to the protocol. The purity and concentration of the extracted DNA were checked using a spectrophotometer. The DNA was then subjected to Conventional PCR for housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, Nested PCR (16S like rRNA and PCR-RFLP (18S rRNA) as described previously and mentioned in Table and Figure 1.8,11,12 The amplified nested PCR product was purified using a QIAquick® PCR purification kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Unidirectional sequencing of the PCR product was performed using an ABI 3730 DNA analyzer. The NCBI-BLAST program was employed to assess the degree of sequence similarity, and the Neighbor-Joining statistical method was applied to construct a phylogenetic tree based on the nucleotide sequences using MEGA-X software version 11.0.13. The DNA of E. moshkovskii was detected using both nested PCR and PCR-RFLP techniques. The SSU rRNA gene was successfully sequenced, and analysis using NCBI BLAST showed a high degree of sequence similarity (>95%) with other E. moshkovskii sequences in the database. Phylogenetic analysis demonstrated that the isolated strain clustered with the reference sequences obtained from NCBI (Figure 2).
Table:
List of primers used in the study
Technique |
Gene |
Primer |
Product Size (bp) |
Ref. |
|---|---|---|---|---|
House Keeping |
GAPDH |
F 5’-GAAGGTGAAGGTCGGAGTCAAC-3’
R 5’-CAGAGTTAAAAGCAGCCCTGGT-3’ |
380 bp |
11 |
Nested PCR |
16S like rRNA-Genus specific Species specific |
EF 1 5’-TAAGATGCACGAGAGC GAAA- 3’ ER 2 5’-GTACAAAGGGCAGGGACGTA3’ EmF 5’-GAAACCAAGAGTTTCAC AAC-3’ EmR 5’-CAATATAAGGCTTGGATGAT-3’ |
553 bp |
12 |
PCR-RFLP |
18S rRNA |
F 5’-AAAGACCAAGTAGGATGAAACTGC-3′
R 5’-TTCCTTCTACTGTTCGGTCTTG-3′ |
1191 bp 1066 bp & 66/59 bp |
8 |
Figure 1. Representative image of 1.8% agarose gel electrophoresis 37 (A). Conventional PCR for housekeeping gene; (B). Nested PCR for E. moshkovskii; (C). PCR-RFLP NC-Negative control; PC-Positive control; S1-Stool sample 1; (D). 2.5% gel image of PCR-RFLP after digestion
Figure 2. Molecular phylogenetic analysis of E. moshkovskii isolate using the SSU rRNA gene by neighbour joining statistical method. The black star represents the E. moshkovskii sequence from the present study while the unmarked sequences represent similar sequences from the Genbank database
Due to the morphological similarities between E. histolytica and other species, earlier studies focused on microscopic identification, which may have contributed to its high reported prevalence. The findings of a recent study from Eastern India reported a decreasing trend in E. histolytica infections in that geographical region. Most of the patients with acute diarrhoea were infected with E. moshkovskii, suggesting that it may be an emerging enteric pathogen in the Indian subcontinent.7 In another study conducted by the same research group, fecal samples from pigs were found to be positive for E. moshkovskii, highlighting the zoonotic potential of the species.13 It is important to note that the primary mode of transmission is through the fecal-oral route. The patient in the present study had a history of consumption of consuming food from outside and poor dietary habits.
There were no other parasites detected in the stool sample as indicated by the results of microscopy apart from E. moshkovskii which was detected via PCR. The patient presented only with abdominal discomfort and altered bowel habits and not diarrhea, a finding similar to the previous study from India.3 There were two more samples which showed a band in PCR corresponding to that of the positive control, but since no sequencing was performed, these are not being described further. However, these patients also presented with similar abdominal symptoms and studies carried out in various regions around the world, including Bangladesh, have reported cases of diarrhoea associated with E. moshkovskii in children aged between 2-5 years.14
In Egypt, the majority of amoebic infections were attributed to non-pathogenic species of Entamoeba.15 Molecular techniques are preferable for distinguishing between Entamoeba species due to their morphological similarities and the limited sensitivity of microscopy, which can lead to potential under-detection of infections.16
The present report, therefore, suggests a pan-India prevalence of Entamoeba moshkovskii. Therefore, in order to determine the current disease incidence from the aforementioned species, more epidemiological studies using molecular approaches from a larger geographic area along with a larger sample size with different patient cohort are needed. This will be helpful in understanding the epidemiology and clinical significance of other Entamoeba species in order to improve the diagnosis and management strategies for amoebiasis.
ACKNOWLEDGMENTS
The authors would like to thank Professor Shinjiro Hamano and Professor Shin-Ichi Inoue, Department of Parasitology, Nagasaki University, Japan for providing us with the positive control of Entamoeba moshkovskii. Authors are also thankful to Department of Science and Technology for providing Devyani Sharma with the fellowship.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RS conceived the study. UD designed the study and provided the samples. DS and DR performed the experiments. DR performed phylogenetic analysis. DS wrote the manuscript. UK supervised and edited the manuscript. RS approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available in the NCBI blast database repository, Accession No.: OQ947379.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Post Graduate Institute of Education and Research (PGIMER) Chandigarh, India, vide letter number INT/IEC/2022/SPL-766 dated 16.04.2022
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Othman N, Ujang JA, Ng YL, Kumarasamy G, Noordin R. Amebiasis. Molecular Advancements in Tropical Diseases Drug Discovery. Academic Press. 2020:1-19.
Crossref - Kawashima A, Yanagawa Y, Shimogawara R, Yagita K, Gatanaga H, Watanabe K. Amebiasis as a sexually transmitted infection: A re-emerging health problem in developed countries. Glob Health Med. 2023;5(6):319-327.
Crossref - Parija SC, Khairnar K. Entamoeba moshkovskii and Entamoeba dispar-associated infections in Pondicherry, India. J Health Popul Nutr. 2005;23(3):292-295.
- Scaglia M, Gatti S, Strosselli M, Grazioli V, Villa MR. Entamoeba moshkovskii (Tshalaia, 1941): morpho-biological characterization of new strains isolated from the environment, and a review of the literature. Ann Parasitol Hum Comp. 1983;58(5):413-422.
Crossref - Clark CG, Diamond LS. Methods for Cultivation of Luminal Parasitic Protists of Clinical Importance. Clin Microbiol Rev. 2002;15(3):329-341.
Crossref - Nath J, Ghosh SK, Bhattacharjee P, Paul J, Singha B. High prevalence of Entamoeba moshkovskii infection in HIV seropositive patients of Barak Valley, Assam, India. BMC Infect Dis. 2014;14(Suppl 3):P1.
Crossref - Sardar SK, Ghosal A, Haldar T, et al. Prevalence and molecular characterization of Entamoeba moshkovskii in diarrheal patients from Eastern India. PLoS neglected tropical diseases. 2023 May 11;17(5):e0011287.
Crossref - Sardar SK, Kobayashi S, Das K, et al. Development of a simple PCR–RFLP technique for detection and differentiation of E. histolytica, E. dispar and E. moshkovskii. Parasitology Research. 2023;122(1):139-44.
Crossref - Singh A, Banerjee T, Khan U, Shukla SK. Epidemiology of clinically relevant Entamoeba spp.(E. moshkovskii /dispar/moshkovskii/bangladeshi): A cross sectional study from North India. PLoS Negl Trop Dis. 2021;15(9):e0009762.
Crossref - Mewara A, Khurana S, Gupta S, Munda VS, Shreya Singh S, Sehgal R. Diagnostic performance of Mini Parasep® solvent-free foecal parasite concentrator for the diagnosis of intestinal parasitic infections. Indian J Med Microbiol. 2019;37(3):381-386.
Crossref - Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21(3):389-395.
Crossref - Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7(1):47.
Crossref - Sardar SK, Das K, Maruf M, et al. Molecular evidence suggests the occurrence of Entamoeba moshkovskii in pigs with zoonotic potential from eastern India. Folia Parasitol. 2022;69:2022.012.
Crossref - Ali IKM, Hossain MB, Roy S, et al. Entamoeba moshkovskii infections in children, Bangladesh. Emerg Infect Dis. 2003;9(5):580-584.
Crossref - Abozahra R, Mokhles M, Baraka K. Prevalence and Molecular Differentiation of Entamoeba histolytica, Entamoeba dispar, Entamoeba moshkovskii, and Entamoeba hartmanni in Egypt. Acta Parasitol. 2020;65(4):929-935.
Crossref - Moran P, Serrano-Vazquez A, Rojas-Velazquez L, et al. Amoebiasis: Advances in Diagnosis, Treatment, Immunology Features and the Interaction with the Intestinal Ecosystem. Int J Mol Sci. 2023;24(14):11755.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.