ISSN: 0973-7510
E-ISSN: 2581-690X
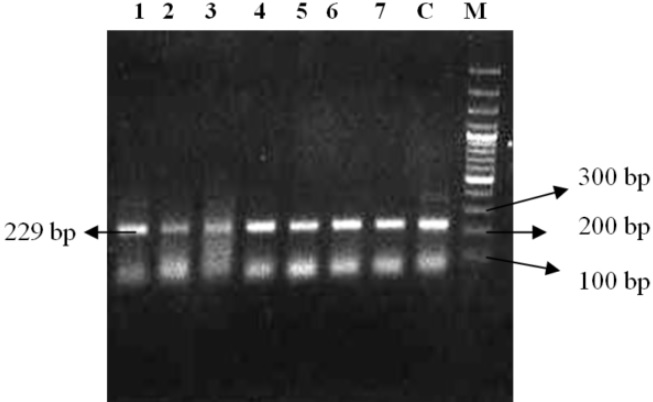
Mycobacterium avium subsp. paratuberculosis is a pathogen that causes johne’s disease in animals and is implicated in Crohn’s disease in humans. Culture of Mycobacterium avium subsp. paratuberculosis (MAP) from faeces has been considered the gold standard for the diagnosis of paratuberculosis for many years. However, direct faecal polymerase chain reaction (PCR) is becoming more widely used, demonstrating similar sensitivity and specificity to culture. In the present study, faecal culture and IS900 Polymerase chain reaction (PCR) assay of faecal samples was done on 200 clinically suspected cases of Johne’s disease in dairy cattle. One isolates appeared only on the mycobactin J supplemented media at 8–16 weeks post-inoculation. A total of 7 faecal samples out of 200 samples were detected positive by IS900 PCR assay for Mycobacterium avium subsp. paratuberculosis (MAP) yielding an expected product of size 229 bp. The sensitivity of the IS900 PCR was assessed by making ten fold serial dilutions of the known concentration (5 ng/µl) of the standard genomic DNA of MAP. The detection limit of the IS900 PCR was upto5 pg/µl.
Mycobacterium avium subsp. paratuberculosis, faecal culture, IS900 PCR.
Paratuberculosis (Johne’s disease) is a chronic enteric progressive disease, characterized clinically by chronic or intermittent diarrhea, emaciation, and death. The disease has worldwide distribution and economic impact on ruminant livestock production (Stabel, 1998). The disease is considered one of the most important diseases of ruminant population (Tripathi et al 2002 and Sivakumar et al 2005). Johne’s disease cause huge economic losses and has high impact on dairy and beef cattle industry due to premature culling of animals, reduced weight gain, reduced feed efficiency, reduced carcass value, reduced milk production, increased susceptibility to mastitis and reproductive disorders leading to increased calving intervals, reduced fertility and additional veterinary costs (Hasonova and Pavlik 2006). Mycobacterium avium subsp. paratuberculosis is biochemically relatively inactive and identification is currently based on mycobactin dependency and detection of the insertion sequence IS900 by the means of the polymerase chain reaction (Collins et al 1989).
Faecal culture may remain the most definitive test to date for identification of both sub-clinically and clinically infected cattle but the prolonged incubation period (12- 16 weeks) required for accurate diagnosis was a distinct disadvantage (Stabel and Whitlock 2001). In addition, the need of a specialized medium for propagation of the organism and frequent contamination is often a problem when culturing MAP from faecal specimens. Despite some of these disadvantages, faecal culture is still considered as the “gold standard” diagnostic test for the detection of Map (Sweeney et al 1992). It is the only test that does not produce false positive results (100% specificity) (OIE 2004). Although the ‘gold standard’ for detection and confirmation of viable or more specifically culturable. MAP remains isolation of the organism in culture, PCR has made rapid detection of this fastidious, slow-growing organism possible in milk, faeces and clinical samples (Grant et al 2000, Pillai and Jayarao 2002 and Fang et al 2002).
The most widely used target gene for detection of MAP IS900 was first described in 1989 (Green et al 1989) and presently considered specific for MAP. The MAP genome has 15 to 20 copies of the insertion element, and the sequenced strain K-10 has 17 copies (Li et al 2005). This high copy number target gives an increased sensitivity compared to systems targeting single copy genes, which makes it popular in molecular diagnostic methods for paratuberculosis. IS900 has a unique nucleotide sequence which can be specifically detected by hybridization or PCR techniques (Collins et al 1993 and Moss et al 1992). Sequences of IS900 proved to be highly sensitive and specific markers of MAP among other slowly growing, acid-fast bacteria, because IS900 has been detected in all reference and vaccine strains as well as in field isolates of MAP from several hosts but never in other bacterial species (Ahrens et al 1995 and Whipple et al 1990).
Since Johne’s disease is so insidious within an animal population, early diagnosis and proper management are considered to be the most useful tools for controlling the disease within domestic livestock herds. Keeping the above points in mind the present study was planned with the objective to detect Mycobacterium avium subsp. paratuberculosis in bovine fecal samples using faecal culture and IS900 PCR assay.
Collection of samples
The present study was carried out to diagnose paratuberculosis using faecal culture and PCR assay on the DNA extracted from faecal samples of cattle and buffaloes suspected for infection with Mycobacterium avium subsp. paratuberculosis on the basis of history of chronic intermittent diarrhoea, emaciation and cachectic condition. Faecal (n=200) samples from cattle and buffaloes with a history of chronic intermittent diarrhoea were collected from dairy farms in Ludhiana, Punjab.
Laboratory Techniques
Isolation of MAP from faecal samples
Attempts were made to isolate Mycobacterium avium subsp. paratuberculosis (MAP) by inoculation of the AFB positive faecal samples on Middlebrook 7H10 media supplemented with mycobactin J (1¼g/ml) (OIE Terrestrial Manual 2008) after following appropriate decontamination procedure by Merkal and Richards (1972). The cultures were incubated at 42° C and were observed periodically for a period of 6 months post inoculation and the growth observed was confirmed by IS900PCR using phenol-chloroform method of DNA extraction (van Embden et al 1993).
DNA Extraction
The extraction of genomic DNA from faecal samples was done using commercially available bacterial genomic DNA Isolation Kit (HiPurA DNA Purification kit) as per the manufacturer’s instructions.
IS900 PCR Protocol
DNA was amplified by PCR using Primers based on IS900 sequence (Table I). PCR was performed as per Vary et al (1990) with some modifications. For the amplification of the IS900 sequence PCR reaction mixture was made in a final volume of 25 µl containing ready to use GoTaq® Green Master Mix, (Promega), forward primer (10 pmol), reverse primer (10 pmol), nuclease free water and DNA template. Cycling conditions were initial denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 45 sec, annealing of primers at 56°C for 45 sec, extension at 72°C for 45 sec and final extension at 72°C for 10 min. PCR products were run by agarose gel electrophoresis and visualized in Gel Documentation System (Alpha Innotech)
Table (1):
Sequences of primers used in IS900 PCR for detection of MAP (Vary et al 1990).
| Primers | Sequence | Size of PCR product |
|---|---|---|
| Forward (IS900/150C) | 5’- CCG CTA ATT GAG AGA TGC GAT TGG – 3’ | 229bp |
| Reverse (IS900/921) | 5’- AAT CAA CTC CAG CAG CAG CGC GGC CTC G -3’ |
Sensitivity of IS900 PCR
The sensitivity of the IS900 PCR was assessed by making a ten fold serial dilutions of the known concentration (5 ng/¼l) of the standard genomic DNA of MAP. The detection limit of the IS900 was upto5 pg/¼l (Figure 1).
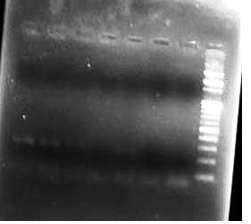 Fig. 1: Sensitivity of PCR using IS900 sequence of MAP
Fig. 1: Sensitivity of PCR using IS900 sequence of MAPIn the present study, 200 faecal samples were processed for the isolation of MAP. After 5 months of incubation, only one faecal sample was found positive for faecal culture (Figure 2 and 3). The animal positive for faecal culture was high degree (4+) positive by ZN staining. The sample was confirmed positive by conventional PCR of the DNA extracted from the colonies (Figure 4). IS900 PCR was also employed directly on the DNA extracted from faecal samples. A total of seven faecal samples (4.29 percent) were detected positive (Figure 5) out of all the 200 faecal samples processed yielding an expected PCR product size of 229 bp.
 Fig. 2. Small raised slightly white colony of MAP
Fig. 2. Small raised slightly white colony of MAP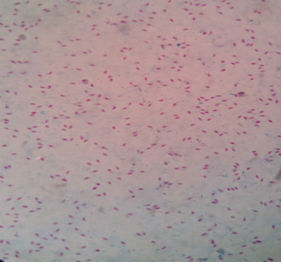 Fig. 3. Acid-fast positive MAP bacilli
Fig. 3. Acid-fast positive MAP bacilli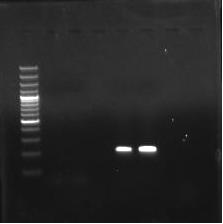 M- 100 bp marker
M- 100 bp markerL1- 229 bp band of isolate
L2- Control
Fig. 4. Confirmation of the MAP isolate by IS900 PCR
The cultural isolation of MAP from faecal samples is considered as the gold standard (OIE, 1996) for diagnosis of MAP. However, culture, though considered more sensitive than most other detection techniques for MAP, is a slow and exacting procedure because of the long generation time of MAP organisms which may extend well beyond 12-16 weeks. Also, the decontamination procedures followed during the processing of faecal samples for culture inoculation also substantially decrease the number of viable MAP bacteria (Whittington and Sergeant 2001). In addition, contamination of culture by overgrowth of competing bacteria and fungal agents, chemical decontaminants used in isolation of MAP, intermittent shedding of acid-fast bacilli by subclinically affected animal and drying of the inoculated media due to long incubation periods is also a problem when culturing MAP from faecal specimens. All these factors make isolation of MAP colonies by culture techniques difficult with lower viable MAP recovery and lesser number of culture positive animals (Grant et al 2005 and Ruzante et al 2006).
All the faecal samples detected positive for MAP by IS900 PCR were from ‘3+’ or ‘4+’ ZN positive animals. The detection limit of direct PCR on faecal samples for paratuberculosis infection is stated to be around 100 organisms per g of faeces. This is similar to the detection limit for culture (Merkal et al 1987). Different types of samples pose different barriers to extraction and PCR. Unfortunately, faecal samples also belong to the samples that are quite difficult to process. It is the method of extraction of mycobacterial DNA from clinical samples rather than the PCR process itself which is the chief bottleneck in detection of MAP by molecular techniques (Englund 2002). PCR is stated as a more sensitive method than culture for detection of Mycobacteria as low levels of MAP shed in the faeces may not be detected by culture and PCR allows us to detect viable as well as non-viable microorganisms (Stabel et al 2002).
- Ahrens, P., Giese, S.B., Klausen, J., Inglis, N.F. Two markers, IS901-IS902 and p40, identified by PCR and by using monoclonal antibodies in Mycobacterium avium strains. J. Clini. Microbiol., 1995; 33:1049-53.
- Collins, D.M., Gabric, D.M., De Lisle, G.W. Identification of repetitive DNA sequence specific to Mycobacterium paratuberculosis. FEMS. Microbiol. Lett., 1989; 60: 175-78.
- Collins, D.M., Hilbink, F., West, D.M., Hosie, B.D., Cooke, M.M., De Lisle, G.W. Investigation of Mycobacterium paratuberculosis in sheep by fecal culture, DNA characterisation and the polymerase chain reaction. Vet. Res., 1993; 133: 599-600.
- Englund, S. Molecular Biology Techniques as a Tool for Detection and Characterization of Mycobacterium avium subsp. Paratuberculosis. Doctoral Dissertation. Department of Bacteriology, National Veterinary Institute, Uppsala, Sweden, 2002.
- Fang, Y., Wu, W.H., Pepper, J.L., Larsen, J.L., Marras, S.A., Nelson, E.A., Epperson, W.B., Christopher-Hennings, J. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine faecal samples. J. Clin. Microbiol., 2002; 40: 287-91.
- Grant, I.R., Hitchings, E.I., McCartney, A., Ferguson, F., Rowe, M.T. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows’ milk. Appl. Environ. Microbiol., 2002; 68: 602-07.
- Grant, I.R., Williams, A.G., Rowe, M.T., Muir, D.D. Efficacy of various pasteurization time-temperature conditions in combination with homogenization on inactivation of Mycobacterium avium subsp. paratuberculosis in milk. Appl. Environ. Microbiol., 2005; 71: 2853-61.
- Hasonova, L., Pavlik, I. Economic impact of paratuberculosis in dairy cattle herds: a review. Vet. Med., 2006; 51(5): 193–211.
- Li, L., Bannantine, J.P, Zhang, Q., Amonsin, A., May, B.J, Alt, D., Banerji, N., Kanjilal, S., Kapur, V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. PNAS USA., 2005; 102: 12344-49.
- Merkal, R.S., Richards, W.D. Inhibition of fungal growth in the cultural isolation of mycobacteria. Appl. Microbiol., 1972; 24(2): 205-7.
- Merkal, R.S., Whipple, D.L., Sacks, J.M., Snyder, G.R. Prevalence of Mycobacterium paratuberculosisin ileocecal lymph nodes of cattle culled in the United States. J. Am. Vet. Med. Assoc., 1987; 190: 676-80.
- Moss, M.T., Sanderson, J.D., Tizard, M.L., Hermon-Taylor, J., El Zaatari, F.A., Markesich, D.C., Graham, D.Y. Polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum in long term cultures from Crohn’s disease and control tissues. Gut., 1992; 33: 1209- 13.
- Office International Des Epizooties. Paratuberculosis (Johne’s disease). Section3.1, Chapter 3.1.6, OIE, Paris, France, 1996.
- OIE Manual. Manual of standards for diagnostic test and vaccines: Paratuberculosis (Johne’s disease) Section 2.2, Chapter 2.2.6, OIE, Paris, France, 2004.
- OIE Terrestrial Manual. Paratuberculosis (Johne’s disease) Section 2.1, Chapter 2.1.11, OIE, Paris, France, 2008.
- Pillai, S.R., Jayarao, B.M. Application of IS900 PCR for detection of Mycobacterium avium subsp. paratuberculosis directly from raw milk. J. Dairy. Sci., 2002; 85: 1052-57.
- Ruzante, J.M., Smith, W.L., Gardner, I.A., Thornton, C.G., Cullor, J.S. Modified culture protocol for isolation of Mycobacterium avium subsp. paratuberculosis from raw milk. Foodborne. Pathog. Dis., 2006; 3: 457-60.
- Sivakumar, P., Tripathi, B.N., Singh, N. Detection of Mycobacterium avium subsp. paratuberculosis in intestinal and lymph node tissues of water buffaloes (Bubalus bubalis) by PCR and bacterial culture. Vet. Microbiol., 2005; 108(3–4): 263–270.
- Stabel, J.R., Whitlock, R.H. An evaluation of a modified interferon– ³ assay for the detection of paratuberculosis in dairy herds. Vet. Immunol. and Immunopathol., 2001; 79(1–2): 69–81.
- Stabel, J.R., Wells, S.J., Wagner, B.A. Relationships BetweenFecal Culture, ELISA, and Bulk Tank Milk Test Results for Johne’s Disease in US Dairy Herds. J. Dairy. Sci., 2002; 85: 525-31.
- Stabel, J.R. Johne’s disease: a hidden threat. J. Diary. Sci., 1998; 81: 283-88.
- Sweeney, R.W., Whitlock, R.H., Hamir, A.N., Rosenberger, A.E., Herr, S.A. Isolation of Mycobacterium paratuberculosis after oral inoculation in uninfected cattle. Am. J. Vet. Res., 1992; 53:1312-14.
- Tripathi, B.N., Munjal, S.K., Paliwal, O.P. An overview of paratuberculosis in animals. Indian J. Vet. Pathol., 2002; 26: 1–10.
- van Embden, J.D.A., Cave, D., Crawford, J.T., Dale, J.W., Eisenach, K.D., Gicquel, B., Hermans, P., Martin, C., McAdam, R., Shinnick, T.M., Small, P.M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J.Clin. Microbiol., 1993; 31: 406.
- Vary, P.H., Anderson, P.R., Green, E., Taylor, J.H., McFadden, J.J. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s disease. J. Clin. Microbiol., 1990; 28(5): 933-37.
- Whipple, D.L., Kapke, P., Vary, C. Identification of restriction fragment length polymorphisms in DNA from Mycobacterium paratuberculosis. J. Clin. Microbiol., 1990; 28: 2561-64.
- Whittington, R.J., Sergeant, E.S. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J., 2001; 79: 387-98.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.