ISSN: 0973-7510
E-ISSN: 2581-690X
Endophytes represent a diverse domain of microorganisms with immense biotechnological potentialities. Metabolites from endophytes, especially fungi, are useful in industrial as well as pharmaceutical aspects. Here, endophytic fungal isolates of Andrographis paniculata collected from the Tapobon, Junglemahal region of West Bengal, India, have been studied for their hydrolyzing enzyme production abilities. Out of eighty-one isolates, twenty-one were positive for amylase, protease, lipase, laccase, and tannase action. Microscopic features of positive isolates revealed that the enzyme producers were Aspergillus sp., Fusarium sp., Alternaria sp., Trichoderma sp., Exerohilum sp., Nigrospora sp., Curvularia sp., Cladosporium sp., Cochliobolus sp., Tricothecium sp., Penicillium sp., Verticillium sp., and Cephalosporium sp. The amylolytic activity was remarkable in the case of endophytic Aspergillus sp. and Fusarium sp. Also, Aspergillus sp. and Alternaria sp. had proteolytic activity. Aspergillus sp., Mycelia sterilia-2, and Trichoderma sp. were potent lipase producers. Along with Exerohilum sp. endophytic Aspergillus sp. had positive laccase activity. The tannic acid degrading activity was highest for Aspergillus sp. APL11 followed by Helicosporium sp. and Fusarium sp. Tannase producing ability of Aspergillus sp. was optimized and an incubation time- 96h, incubation temperature- 29°C, initial medium pH- 6.5, carbon source- glucose, and nitrogen source- NaNO3 exhibited a two-fold scale-up of (2.83±0.05 u mL-1) in tannase action. These enzymes offer eco-friendly and efficient solutions across a wide range of industrial processes including food production, textile processing, and pharmaceutical synthesis. Fungal endophytes of medicinal plants act as an alternative source for hydrolysing enzymes.
Hydrolysing Enzyme, Fungal Endophytes, Optimization, Tannase
Endophytic fungi hold numerous biotechnological applications and have been exploited in recent decades for the discovery of novel metabolites with pharmaceutical applications. They provide protection to their host and perform multidisciplinary functions, i.e. render resistance to biotic-abiotic stress situations (water, salinity, and temperature), modify the hosts’ physiological properties, synthesise different phytohormones, enzymes, etc. Endophytic fungi are favoured for enzyme production over endophytic bacteria for their easy identification, mass production, economically feasible media requirements, and consistency in production. Endophyte biology has caught attention at a later stage in the last decade but now is being explored logarithmically for the production of industrially relevant molecules.1 The endophytes of ethnomedicinal plants represent a pool of novel metabolites with a wide array of biodiversity. The exploration of endophytes and their enzyme production has opened up new avenues in the field of biotechnology. They offer a rich source of enzymes with diverse applications in industries such as pharmaceuticals, food processing, and environmental remediation. The potential of endophytes as enzyme producers is particularly promising, given their close association with plants and the unique biochemical pathways they possess. By tapping into this unexplored area, researchers are actively working towards harnessing the vast potential of endophytes for various industrial applications. Among the array of enzymes amylases, proteases, tannases, lipases, and laccases, stand out as potential game-changers in various industrial sectors. As researchers continue to unravel the mysteries of endophytes it may redefine the landscape of the enzyme industry forever. Amylase, the oldest known enzyme, accounts for 30 % of total enzymes exploited industrially and it breaks down starch by targeting α-1,4 glycosidic bonds.2 Endophytes produce this enzyme to utilise amylose as the carbon source. These are used for starch-to-sugar syrup conversion, cyclodextrin production, etc.3 Protease is a type of hydrolysing enzyme that cleaves the peptide bonds on a polypeptide chain and forms a small fraction of peptides. They are used in numerous industrial sectors like baking (biscuit manufacturing), brewing, textiles, and pharmaceuticals in areas such as oncology, inflammatory regulations, blood rheology, and immune regulation.4 Lipases are lipid-degrading water-soluble acyl hydrolases, that are widely distributed among all life forms. They also hold pharmaceutical utilities. Whereas laccases, also known as benzenediol: oxygen oxidoreductase (EC 1.10.3.2), are multi-copper oxidases that facilitate the oxidation of diverse substrates- diphenols, methoxy-substituted monophenols, aliphatic, aromatic amines and are efficient agents of bio-remediation. Tannase is industrially valuable for its use in pharmaceuticals, food, and beverages. It was earlier isolated from leaves, stems, fruit, bark, and branches of the plant and has immense usage in beverage industries. The definition of endophytes reveals that they reside within the host plant so there is a huge chance that endophytes could be potent tannase producers and also they promote plants’ adaptation in new environments.5
In the present investigation fungal endophytes of a common ethnomedicinal plant Andrographis paniculata have been screened for the production of industrially valuable extracellular hydrolytic enzymes, i.e. protease, lipase, amylase, laccase and tannase. They were first screened in solid media and then further evaluated in respective broths. The potent isolates were identified based on macroscopic morphology and molecular characteristics. Isolate APL11 was the most potent tannase producer and was identified as Aspergillus flavus. Tannase production was optimized through One Variable at A Time and Response Surface Methodology techniques. The produced tannase was also biochemically characterised and was found to be more efficient than contemporary ones. The present investigations elucidate the rich diversity of endophytic fungi of ethnomedicinal pants and shed light on the industrial applicability of fungal endophytes as potent producers of pharmaceutically valuable enzymes.
Plant materials collection and endophytes isolation
The undamaged and disease-free plant specimens were gathered and carefully sealed within zip-lock polythene bags in Tapobon, Paschim Medinipur, West Bengal, India. The region is characterized by a tropical, warm, and humid climate, featuring an average temperature of 34°C. Tapobon is situated between latitudes 22°25′ to 22°57′ North and longitudes 87°11′ East, with an altitude of 23 meters. The primary plant parts utilized for extracting endophytic fungi were the leaves and stems. To eradicate the epiphytic microbes surface sterilization was done using sodium hypochlorite and 70% alcohol. In water agar plates the thin (1.0 cm x 1.0 cm) segments of the sterile samples were placed and incubated at 25°C in a BOD incubator. Within 72-96 hours of incubation, fungal hyphae from most of the samples emerged which were then transferred to PDA (potato dextrose agar) plates.
Identification of isolated endophytes
After fifteen days of incubation, endophytic fungi were characterized through an assessment of their visible and internal features and examined under a light compound microscope (Leica DM 3000). Fungal genera were identified by macroscopic and microscopic morphology. Each of the isolated and recognized endophytic fungi was designated with unique codes and used for further studies. Isolate APL11 was subjected to molecular identification following the standard protocols and the obtained nucleotide sequence was submitted to GenBank. A phylogenetic tree compared with other isolates was prepared for the identification of APL11.6 Calculation of the colonization frequency of each isolate, in addition to assessing diversity using indices such as Shannon–Wiener, Simpson’s dominance, Simpson’s diversity, and species richness. These calculations were performed in accordance with established methodologies as outlined in Santra and Banerjee.7
Assay for the production of extracellular enzymes
The screening process involved two methods: a qualitative approach, known as the agar plate method, and a quantitative method, referred to as the liquid culture method. To assess the functional role of extracellular enzymes produced by fungal endophytes, the fungi were cultivated on potato dextrose agar (PDA) for a period of 6-7 days, with incubation at 25°C. Subsequently, 5 mm mycelial plugs were placed on solid media. Following this incubation, at room temperature, the extent of enzyme activity surrounding the fungal colony was measured, as described by Krishnan et al. in 2016.8 The procedure used for both the qualitative and quantitative estimation of amylolytic, proteolytic, lipase, laccase, and tannase activity is detailed below.
Screening of fungal endophytes for amylase production and estimation of amylase activity
For the qualitative analysis of amylase production, fungal isolates were cultivated on GYP (Glucose Yeast Extract Peptone Agar) medium containing soluble starch at a concentration of 0.2% and adjusted to pH 6.0. Following 72 hours of incubation, the plates were flooded with a solution composed of 1% iodine and 2% potassium iodide. The presence of a clear zone surrounding the fungal colonies served as an indicator of amylolytic activity. For quantitative analysis, the isolates that exhibited positive outcomes in the iodine test on solid media were chosen. These selected isolates were cultured in 80 mL of basal media within Erlenmeyer flasks, which were supplemented with 1.5% starch. Flasks were sterilized and cooled, and 5 mm2 PDA plugs containing fungal mycelia were inoculated into each flask. An uninoculated flask with the same media composition served as the control. Liquid samples were collected from the flasks every 24 hours for a period of 5 days in 2 mL Eppendorf tubes, starting from 48 hours of incubation
Completing the incubation phase, the broth was first filtered using Whatman No. 1 filter paper and subsequently subjected to centrifugation at 5000 revolutions per minute (rpm) for 15 minutes at a temperature of 40°C. The supernatant, which contained liberated reducing sugars, was used for further analysis. The quantification of reducing sugars was performed utilizing the DNS (dinitrosalicylic acid) method.9
To determine the amylase activity, a reaction mixture was prepared using 1 mL of 0.2 molar phosphate buffer (K2HPO4 + KH2PO4) with a pH of 6.0, 1% starch solution 0.5 mL (w/v) and the crude enzyme extract 0.5 mL. The reaction mixture was then incubated at 37°C for approximately 45 minutes. To stop the reaction, 2 mL of DNS solution was added to each sample. A separate estimation at 0-time was also performed to determine the initial glucose concentration in the solution before the reaction started. The solutions were boiled in a water bath for 15 minutes, resulting in a reddish-brown color due to the release of reducing sugars. The optical density of solutions was measured at a wavelength of 550 nm, employing a UV-spectrophotometer (Shimadzu Corp., Serial AI-4547-0302).
To quantify the amount of sugars produced in the samples, a standard curve was established using a stock solution of glucose with a concentration of 1 mg/mL at various concentrations (20, 40, 60, 80, 100 µg/mL). The absorbance of the samples was then compared to this standard curve to determine the sugar content. In this context, one unit of amylase activity was defined as the quantity of enzyme that releases 1 mole of reducing sugar, such as maltose or glucose, per minute under the specified assay conditions.9
Screening of fungal endophytes for protease action and estimation of protease production
Protein breaking ability of endophytic fungus was assayed on a modified Czapek Dox medium with a composition of KH2PO4 1.5 g, (NH4)2SO4 0.5 g, MgSO4, 7H20 0.06 g and CaCl2, 2H2O 0.06 g along with separately sterilized 1 g milk powder in a 100 ml of media composition of pH 7.0.10 Mycelia plugs from pure fungal cultures were placed on the plates and incubated for 5 days at 25°C. Positive protease producers were identified according to the halo clear zone surrounding the fungal colony. For quantitative analysis, protease-producing fungal isolates were cultured in 250 mL Erlenmeyer flasks, each filled with 80 mL of basal salt media. The composition of the basal salt media per liter included: NaNO3-2.5 g, KH2PO4 – 1 g, MgSO4·7H2O-0.5 g, and KCl-0.5 g. and obviously supplemented with 1%(w/v) powder milk. The medium was sterilized, cooled, and aseptically inoculated with the selected fungi and incubated for 7 days at 150 rpm. Starting from 48 hours of incubation, the liquid was collected every 24 hours for the remaining 5 days.
The liquid samples were centrifuged at 8000 rpm for 15 minutes at 4°C in a cooling centrifuge (REMI-AACI-9575; C-2432). The resulting supernatant was utilized for assessing proteolytic activity using casein as the substrate. Specifically, 0.5 mL of the supernatant was mixed with 0.5 mL of a 1% (w/v) casein solution in a 100 mM sodium acetate buffer with a pH of 5.5. This mixture was then incubated for 30 minutes at 45°C, and the reaction was stopped by adding 0.5 mL of trichloroacetic acid (TCA). After a 15-minute incubation at room temperature, the mixture was centrifuged at 6000 rpm for 15 minutes to remove debris. The absorbance of the soluble fraction was measured at 650 nm using a UV-spectrophotometer after the addition of Folin-Ciocalteu’s reagent, and the protease activity was quantified based on a standard curve created using tyrosine solutions with concentrations ranging from 20 to 100 mg/L.1 The protease activity was expressed as the amount of enzyme releasing 1 µmol of tyrosine per minute according to the standard curve.
Screening of fungal endophytes for lipase Activity and estimation of lipase production
To assess the production of lipolytic enzymes, fungi from pure culture plates were cultivated on Peptone Agar medium with peptone, NaCl, CaCl2·2H2O, and agar at a pH of 6.0. This medium was enriched with separately sterilized Tween 20 at a concentration of 1%. A 5 mm2 fungal plug was inoculated onto the Peptone Agar plates and incubated for 5 days. At the end of this incubation period, the presence of lipase activity was confirmed by the formation of a visible precipitate around the fungal colony. This precipitate results from the conversion of lauric acid into its calcium salt, which is released by the lipase enzyme. For the quantitative determination of lipase enzyme activity, fungal samples were cultivated in 250 mL flasks, each containing 100 mL of autoclaved medium. This medium included (NH4)2SO4, MgSO4, NaCl, Ca(NO3)2, KH2PO4, glucose, and yeast extract, with the pH adjusted to 5.5. Sterilized olive oil was separately added at a rate of 1 mL per 100 mL of sterile medium. The samples were incubated at 30°C and agitated at 110 rpm on a BOD shaker for 4 days. Every 24 hours, 2 mL of samples were collected and employed for the enzyme assay, as described by Geoffry and Schur.11
The collected samples underwent centrifugation at 5000 rpm for 20 minutes, and the resulting supernatant was employed to assess lipase activity. For the lipase assay, an emulsifying agent was created by combining 0.5 mL of olive oil, 0.5 mL of phosphate buffer (0.066M at pH 7), and 0.5 mL of tween 20 in equal proportions. In the lipase assay, 1 mL of the emulsifying agent was mixed with 1 mL of the supernatant and shaken for 3 minutes. This mixture was then placed in an incubator shaker at 37°C and 150 rpm for 5 hours. Following incubation, the optical density (OD) was measured spectrophotometrically at a wavelength of 655 nm. Lipase activity was defined as the amount of enzyme capable of releasing 1µmol of fatty acid at 37°C and pH 7 under the specified assay conditions.11
Screening of fungal endophytes for tannase action and estimation of tannase production
To detect tannase-producing endophytic fungal strains, a plate assay was conducted using the approach outlined in the methodology by Roy et al.12 Modified Czapek Dox medium was prepared with specific compositions of various ingredients. Separate solutions of 1% tannic acid and the prepared medium were sterilized and mixed under laminar airflow conditions. Plugs of mycelia measuring 5mm2 from each fungus were introduced onto Petri dishes filled with the appropriate medium, followed by an incubation period of 6 days at 27°C. The observation of a distinct clear halo surrounding the fungal colony served as confirmation of tannic acid breakdown. For tannase production, 100mL of modified liquid Czapek Dox medium was used, containing tannic acid (1g), NH4Cl (0.3g), CaCl2 (0.05g), NaH2PO4 (0.05g), Na2HPO4 (0.05g), MgSO4 (0.1g), and yeast extract (0.05g) The media were sterilized in an autoclave and then cooled to room temperature. Mycelia plugs from PDA (Potato Dextrose Agar) were transferred to the liquid media, and the inoculated media were placed in a shaker BOD incubator at 150 rpm for 3 days.
To estimate the tannase activity, the collected liquid containing the crude enzyme was used. The remaining tannic acid was quantified by performing the Bovine Serum Albumin (BSA) precipitation method described by Mahapatra et al.13 The following solutions were prepared: a 0.2 M phosphate buffer with a pH of 5.5, a Bovine Serum Albumin (BSA) solution at a concentration of 1 mg/mL, an SDS-triethanolamine solution (containing 1% w/v SDS in 5% v/v triethanolamine), and a 0.13 M FeCl3 solution. The tannase reaction was initiated by combining 0.1 mL of the enzyme solution with 0.3 mL of a 1% (w/v) tannic acid solution in the 0.2 M phosphate buffer (pH 5.5). The reaction mixture was subsequently incubated at 40°C for duration of 30 minutes. Control estimation without an enzyme was also prepared. To detect the initial tannic acid concentration before the start of the reaction, a 0-time estimation was performed.
Following the incubation, the reaction was halted by introducing 2 mL of Bovine Serum Albumin (BSA) at 0°C. The mixture was subsequently centrifuged at 5000 rpm for 10 minutes, and the supernatant was removed. The precipitate was re-suspended in 2 mL of the SDS-triethanolamine solution. To this resuspended solution, 1 mL of FeCl3 (0.13 M) was added, and the absorbance of the resulting solution was gauged at 550 nm using a spectrophotometer.
Screening of fungal endophytes for laccase production and estimation of laccase activity
To determine laccase activity in endophytic fungi, they were cultivated on GYP agar medium supplemented with 0.05% α-Napthol. The medium composition consisted of peptone (10 g/L), yeast extract (5 g/L), glucose (20 g/L), and agar (15 g/L) at pH 7.0. The presence of laccase was confirmed by a noticeable color change in the media, shifting from colorless to brownish-violet, signifying the oxidation of colourless α-napthol by the endophytic fungi, following the method described by Umar and Ahmed, 2022. For the quantitative assessment of enzyme activities, Czapex Dox medium was employed as the liquid fermentation medium. A 5 mm² disc of actively growing fungal mycelia from the periphery was aseptically inoculated into the broth. The culture was then incubated at 25°C for 7 days. The liquid was collected and stored at 4°C for subsequent enzymatic assays.
Following the completion of the incubation period, the collected liquids were subjected to centrifugation at 5000 rpm for 15 minutes at 4°C. The resulting supernatant was utilized to assess laccase activity, which was quantified in U/mL. To eliminate the potential influence of hydrogen peroxide (H2O2) produced by the fungus, catalase (1,000 U/mL) was introduced to the assay solution and allowed to incubate for 1 hour at 37°C.14
Laccase activity was performed spectrophotometrically at 420 nm, using 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulfonate) (ABTS) as a substrate. The assay mixture consisted of 150 µL of 0.5 mM ABTS substrate, 2.7 mL of 0.1 M sodium acetate buffer at pH 4.5, and 150 µL of the culture supernatant. The mixture was incubated for 5 minutes, after which the absorbance was measured at 420 nm using a spectrophotometer, referencing an appropriate enzyme and substrate blank. Laccase activity was defined as the quantity of enzyme capable of oxidizing 1 µmol of ABTS per minute, and these activities were expressed in U/mL. All assays were conducted in triplicate.
Identification of the fungal endophytes
Hydrolysing enzymes perform a crucial role in various biological, industrial, and environmental events by breaking down complex molecules through hydrolysis. They are essential for digestion, cellular metabolism, detoxification, waste decomposition, and in a wide range of industrial applications, including food processing, detergent manufacturing, and environmental remediation. Specific enzymes like amylase, protease, lipase, tannase, and laccase have diverse industrial uses, contributing to improved product quality and environmental stewardship. In total, one hundred and sixty-five endophytic fungal taxa were successfully retrieved from the two hundred and twenty-three samples (explants) of leaf tissues. The colonization frequencies of the fifteen different isolates are documented in Supplementary Table 1, with Aspergillus sp. exhibiting the highest frequency at 8.96 (as seen in Supplementary Table 1). The diversity of the endophytic community within the leaf tissues is presented in tabular format (Supplementary Table 2). The outcomes derived from diversity indices suggest a commendable range of endophytic fungal diversity within the leaf tissues of the explants.
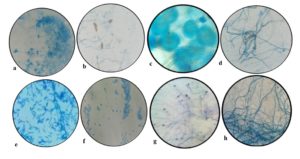
Here, in total twenty-two endophytic fungal isolates were screened as potent enzyme producers. The isolates are characterized through an assessment of their visible and internal morphologies (Figure 1). The most potent tannase producer APL11 was identified as Aspergillus flavus with a GenBank accession number of OR689609 (Supplementary Figure 1). Farouk et al. documented the amylase action of fungal endophytes- Aspergillus, Fusarium, Trichoderma, and dark sterile mycelia.9 A recent investigation from Hawar (2022) elucidated the protease, lipase and laccase-producing abilities of fungal endophytes- Aspergillus flavus, Aspergillus fumigatus, Aspergillus niger, Cladosporium sp., Rhizopus sp., and Mucor sp. of Ziziphus spina.1 Malubag and his coworker found that endophytic fungi F. solani associated with bananas (Musa paradisiaca L.) exhibits amylase and laccase action.15 Current research also supports the previous findings. Enzymes produced by endophytic fungi are particularly valuable due to their stability, substrate specificity, and environment-friendly cultivation practices. They serve as a rich source of biocatalysts with diverse characteristics, making them valuable for research and development across industries.
Figure 1. Endophytic fungal isolates of Andrographis paniculata a- Penicillium sp., b- Curvularia sp., c- Aspergillus sp., d- Cephalosporium sp., e- Fusarium sp., f- Alternaria sp., g- Nigrospora sp., h- Cladosporium sp.
Production of hydrolyzing enzymes
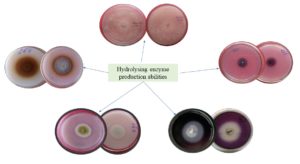
Isolates were tested for their five selected types of hydrolysing enzyme production ability (Supplementary Figure 2). The amylolytic, proteolytic, and lipolytic activity were tested in the solid plate method (Table 1 and Figure 2) and the potent isolates were further screened in liquid media for their quantitative assessment (Table 2). Out of the identified twelve endophytic isolates Aspergillus sp., Fusarium sp., and Trichoderma sp. showed the highest amylolytic activity. The isolate Trichoderma sp. showed the best amylolytic activity which may break the a or b- 1,4 glycosidic bond to utilize the starch provided in the media in its small proportion into glucose or maltose. As amylase is the highest used digestive enzyme, the purified form can be utilized in industries related to the same. Endophyte Trichoderma sp. has not been subjected to amylase production earlier.
Table (1):
Screening of fungal endophytes through Solid Plate Assay (SPA) to produce five selected hydrolysing enzymes
Name of the isolates |
Amylase |
Protease |
Lipase |
Tannase |
Laccase |
|---|---|---|---|---|---|
Trichoderma sp. |
+ |
_ |
+ |
_ |
_ |
Trichothecium sp. |
+ |
+ |
+ |
_ |
+ |
Sterile Mycelia-1 |
+ |
+ |
+ |
_ |
_ |
Aspergillus sp. |
+++ |
+++ |
+++ |
+++ |
+++ |
Sterile Mycelia-2 |
– |
+ |
++ |
– |
– |
Verticillium sp. |
+ |
+ |
_ |
_ |
+ |
Cephalosporium sp. |
+ |
+ |
+ |
_ |
_ |
Fusarium sp. |
++ |
+ |
+ |
+ |
+ |
Alternaria sp. |
+ |
++ |
_ |
_ |
_ |
Penicillium sp. |
+ |
_ |
+ |
+ |
_ |
Helicosporium sp. |
_ |
_ |
+ |
++ |
_ |
Nigrospora sp. |
_ |
_ |
_ |
+ |
_ |
Exerohilum sp. |
_ |
_ |
_ |
+ |
++ |
Unidentified 1 |
_ |
_ |
_ |
+ |
_ |
Unidentified 2 |
_ |
_ |
_ |
+ |
_ |
Table (2):
Different types of hydrolysing enzyme producing potentialities of fungal endophytes of Andrographis paniculata in Liquid Broth Assay (LBA)
| Name of the isolates | Enzymatic activity (U mL-1) | ||||
|---|---|---|---|---|---|
| Amylase | Protease | Lipase | Tannase | Laccase | |
| Trichoderma sp. | 0.81±0.03 | – | 0.63±0.04 | – | – |
| Trichothecium sp. | 0.62±0.02 | 0.73±0.04 | 0.17±0.009 | – | 0.69±0.05 |
| Sterile Mycelia-1 | 0.54±0.02 | 0.61±0.05 | 0.30±0.02 | – | – |
| Aspergillus sp. | 1.19±0.04 | 1.11±0.06 | 0.97±0.06 | 1.77±0.11 | 1.05±0.06 |
| Alternaria sp. | 0.38±0.01 | 0.99±0.03 | – | – | – |
| Verticillium sp. | 0.27±0.02 | 0.47±0.02 | – | – | 0.81±0.04 |
| Cephalosporium sp. | 0.59±0.03 | 0.41±0.03 | 0.29±0.02 | – | – |
| Fusarium sp. | 0.93±0.04 | 0.79±0.04 | 0.51±0.04 | 0.91±0.05 | 0.47±0.03 |
| Sterile Mycelia-2 | – | 0.57±0.03 | 0.77±0.05 | – | – |
| Penicillium sp. | 0.41±0.03 | – | 0.34±0.03 | 0.63±0.04 | – |
| Helicosporium sp. | – | – | – | 1.09±0.09 | – |
| Nigrospora sp. | – | – | – | 0.86±0.08 | – |
| Exerohilum sp. | – | – | – | 0.35±0.03 | 0.83±0.05 |
| Unidentified 1 | – | – | – | 0.57±0.04 | – |
| Unidentified 2 | – | – | – | 0.69±0.06 | – |
Figure 2. Hydrolysing enzyme producing potentialities of fungal endophytes a- amylolytic activity of Fusarium sp., b- proteolytic activity of Aspergillus sp., c- lipase action by Trichoderma sp., d- tannase action by Helicosporium sp., e- laccase activity by Exerohilum sp.
Aspergillus sp. was a potent protease producer along with Alternaria sp. The lipid-breaking activity was found to be remarkably higher in the case of Aspergillus sp. and Trichoderma sp. Also, Aspergillus sp. and Helicosporium sp. were potent tannase producers in comparison to other isolates. Laccase activity was highest in the case of Aspergillus sp. but endophytic Exerohilum sp., Verticillium sp., and Tricothecium sp. were also unique in this respect. The isolates Fusarium sp. and Aspergillus sp. showed the best result in lipase production.16 Lipase has large pharmaceutical and some other industrial applications as it can break the lipid and release fatty acid residues and glycerol.17 The assayed endophytic sources of this enzyme can further be characterized to know the specific activity. Soil fungi are already used in this respect, but endophytes are not well-established. So, more research can be conducted regarding the endophytic lipase producers. Protease can cleave the long polypeptide chains into small amino-acid fragments. Due to their unique properties, they have large applications in industries like detergent, meat tenderisation, etc. The endophytic fungal strain Aspergillus sp. and Alterneria sp. which shows the best result in proteolytic activities could be studied regarding their purification and characterization.18
Tannase or tannic acid hydrolase mainly breaks the tannic acid and releases gallic acid, glycerol esters, and glucose. Gallic acid has great applications in food and pharmaceuticals. The endophytic isolate Aspergillus flavus APL11 shows the highest activity in tannase production in the suitable media supplied with tannic acid which is confirmed by both optical density measurement and HPLC of the liquid samples.19 Further studies regarding the characterization and purification of this enzyme for the said fungal isolate could be conducted.
The enzyme laccase is unique as they have greater utilization in bioremediation and beverage stabilization. This enzyme releases the Cu+ ions by oxidation of α-Napthol. The presence of this enzyme can be detected by supplying the a-Naphthol in media.20 Laccase production by soil fungi has been reported but endophytic fungi are rare in this occasion. The endophytic fungal isolates Exerohyllum sp. and Trichothecium sp. showed promising activity in the present study and can also be further investigated regarding purification and characterization.
Endophytes are the untapped source of knowledge that has not been utilized and exploited to its best till date.21 This is high time to turn to endophytic fungal sources for the search for bioactive compounds. In this study, we have selected the flora of the Tapobon region of Paschim Medinipur district based on the virgin and undisturbed ecological conditions. Soil microorganisms have been tremendously tested to produce industrially valuable enzymes, but endophytic flora has not been widely explored. So, this study could open up new horizons in that respect. Enzymes like amylase, protease, and lipase are of huge pharmaceutical and industrial value. So, the search for a new type of amylolytic, proteolytic, and lipolytic enzymes is justified. Enzymes like arginase and laccase are new bioactive agents that can be exploited in several aspects ranging from mycoremediation, xenobiotics degradation, immunoassays, as linkers in organic synthesis, in cosmetic industries, and finally in the pharmaceutical industry as medicine of Alzheimer’s disease, anti-leishmaniasis, cystic fibrosis, age-related endothelial dysfunction, etc. Research on industrially essential enzymes for instance amylase, protease, lipase, tannase, and laccase is essential for enhancing industrial processes, reducing resource consumption, and promoting sustainability. These enzymes find applications in various sectors, including food processing, textiles, detergents, pharmaceuticals, and more, offering economic value and contributing to cleaner ecosystems through environmental remediation efforts.
Moreover, studying these enzymes can advance biotechnology, lead to cost-effective solutions, open new markets, and even uncover novel enzymes from various sources, fostering innovation and making industries more efficient, sustainable, and competitive. Ultimately, the present research holds the potential for medical and pharmaceutical benefits as well, making it a multifaceted and critical area of exploration for both scientific and industrial advancement.
Optimizations of tannase production
Tannase production from endophytic Aspergillus flavus APL11 was optimized by One Variable At A Time (OVAT). The fermentation conditions were revealed as best suited for tannase production. There was approximately a two-fold increase (Supplementary Figure 3) in tannase activity in post-optimized conditions (Table 3). Meenashree and his associates reported six endophytic fungi from the root tissue of Eichhornia crassipes as positive tannase producers. As per the reports, endophytic Penicillium samsonii and Penicillium minioluteum are tannase producers.22 Aspergillus niger from Tamarindus indica has been reported by D’Souza et al. as a tannase-positive isolate.23 Also, Pakaweerachat reported Aspergillus niger for tannase and gallic acid production by the valorization of tannin-rich triphala waste in solid-state fermentation (SSF).24 All these findings supported the current study on tannase production by endophytic APL11.
Table (3):
Optimization of different types of fermentation parameters for the Tannase production by fungal endophyte Aspergillus sp. APL11 through OVAT approach
Parameters Evaluated |
Critical values |
The concentration of the components (g L -1) |
Tannase activity (U mL-1) |
|---|---|---|---|
Incubation time (h) |
24 |
– |
0.27±0.03 |
48 |
– |
0.51±0.05 |
|
72 |
– |
0.83±0.07 |
|
96 |
– |
1.05±0.02 |
|
120 |
– |
0.96±0.11 |
|
Incubation temperature (°C) |
25 |
– |
0.99±0.07 |
27 |
– |
1.17±0.04 |
|
29 |
– |
1.33±0.05 |
|
31 |
– |
1.24±0.08 |
|
33 |
– |
1.11±0.03 |
|
Initial medium pH |
6 |
– |
1.38±0.09 |
6.5 |
– |
1.46±0.02 |
|
7 |
– |
1.39±0.05 |
|
7.5 |
– |
1.32±0.06 |
|
Additional carbon sources |
Fructose |
0.5 |
1.33±0.03 |
Glucose |
0.5 |
1.69±0.03 |
|
Maltose |
0.5 |
1.29±0.05 |
|
Starch |
0.5 |
1.58±0.05 |
|
Lactose |
0.5 |
1.30±0.09 |
|
Galactose |
0.5 |
1.46±0.10 |
|
Mannitol |
0.5 |
1.23±0.12 |
|
Additional nitrogen sources |
Peptone |
0.5 |
1.53±0.07 |
NaNO3 |
0.5 |
2.07±0.02 |
|
NH4NO3 |
0.5 |
1.87±0.11 |
|
Urea |
0.5 |
1.71±0.09 |
|
NH4Cl |
0.5 |
1.83±0.05 |
|
KNO3 |
0.5 |
1.96±0.05 |
|
Yeast extract |
0.5 |
1.49±0.04 |
|
Beef extract |
0.5 |
1.41±0.03 |
|
Glucose concentration |
Glucose |
0.5 |
2.07±0.04 |
1.0 |
2.35±0.04 |
||
1.5 |
2.14±0.04 |
||
2.0 |
2.03±0.04 |
||
NaNO3 concentration |
NaNO3 |
0.25 |
2.02±0.06 |
0.5 |
2.35±0.06 |
||
0.75 |
2.470±0.04 |
||
1 |
2.58±0.08 |
||
1.25 |
2.23±0.14 |
||
1.5 |
2.19±0.10 |
||
Different metal ions |
MgCl2 |
0.1 |
2.52±0.11 |
FeCl3 |
0.1 |
2.22±0.06 |
|
KCl |
0.1 |
2.83±0.05 |
|
NaCl |
0.1 |
2.28±0.06 |
|
NaH2PO4 |
0.1 |
2.67±0.09 |
|
K2HPO4 |
0.1 |
2.71±0.02 |
|
KH2PO4 |
0.1 |
2.63±0.11 |
Biochemical characterization of the tannase
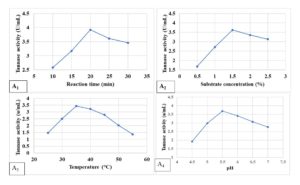
It was seen that the efficiency of the semi-purified enzyme was greatly influenced by different assay conditions like time of incubation, initial medium pH of the reaction mixture, incubation temperature, and concentration of substrate. To achieve the maximum efficiency of tannase, some physicochemical properties of crude tannase were studied. A 20-minute reaction duration (Figure 3A1) was found as the most suited one for the highest tannase activity (3.92 U mL-1). Maximum tannase action (3.62 UmL-1) was detected at 1.5% Tannic acid (substrate) concentration (Figure 3A2). Tannase from endophytic A. flavus APL11 was found to be optimally active at 35±2°C of 3.43 U mL-1 and 60% enzyme activity was retained at the above-said temperature after 2 h of incubation (Figure 3A3). Tannase synthesized by APL11 exhibited the highest gallic acid production at a pH of 5.5 (3.69 UmL-1). Liu et al. reported that tannase from Aspergillus melleus functions at 40°C with a pH of 5.5 whereas tannase from A. niger and A. fumigatus was effective at 30°C, at pH 4.19, 25 Rivas et al. documented that tannase from microbial sources even functions around 20-60°C though in most cases 35°C is the ideal temperature.26 Also, in the case of the most suitable pH tannase has a range of around 5.5 (Figure 3A4).27 Tannase earlier reported from Aspergillus sp. is mostly similar to the presently reported tannase from A. flavus APL11 from their biochemical perspectives.
Hydrolytic enzymes, especially tannase, hold immense utility in the sector of food and pharmaceuticals. Extracellular enzymes from endophytic fungi are a less explored area of research and here we have isolated fungal endophytes from ethnomedicinal plant Andrographis paniculata. Isolates are found to be potent producers of tannase, amylase, protease, lipase, laccase and can be exploited from pharmaceutical and food technology perspectives. The research on ‘tannase and other enzymes’ is proliferating over the century still, there is always an exponential requirement for cost-effective novel and stable enzymes from untapped biological sources. Here, tannase optimization was performed through statistical methods i.e., OVAT and RSM and was characterized biochemically. The optimisation results are somewhat superior to the contemporary tannase-producing isolates. Our report on tannase, and other hydrolysing enzymes from the endophyte of Andrographis paniculata is the first report according to our best knowledge.
Additional file: Additional Table S1-S2 and Figure S1-S3.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Hawar SN. Extracellular enzyme of endophytic fungi isolated from Ziziphus spina leaves as medicinal plant. Int J Biomater. 2022;13597.
Crossref - Patil AG, Khan K, Aishwarya S, et al. Fungal amylases and their industrial applications. Industrially Important Fungi for Sustainable Development: Bioprospecting for Biomolecules. 2021;2:407-434.
Crossref - Burhan A, Nisa U, Gokhan C, Omer C, Ashabil A, Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochem. 2003;38(10):1397-1403.
Crossref - Shankar Naik B, Abrar S, Krishnappa M. Industrially important enzymes from fungal endophytes. Recent Advancement in White Biotechnology Through Fungi: Diversity and Enzymes Perspectives. 2019;1:263-280.
Crossref - Mishra R, Kushveer JS, Revanthbabu P, Sarma VV. Endophytic fungi and their enzymatic potential. Advances in Endophytic Fungal Research: Present Status and Future Challenges. 2019:283-337.
Crossref - Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022-3027.
Crossref - Santra HK, Banerjee D. Broad-spectrum antimicrobial action of cell-free culture extracts and volatile organic compounds produced by endophytic fungi Curvularia eragrostidis. Front Microbiol. 2022;13:920561.
Crossref - Krishnan A, Convey P, Gonzalez-Rocha G, Alias SA. Production of extracellular hydrolase enzymes by fungi from King George Island. Polar Biol. 2016;39(1):65-76.
Crossref - Farouk HM, Attia EZ, El-Katatny MMH. Hydrolytic enzyme production of endophytic fungi isolated from soybean (Glycine max). J Mod Res. 2020;2(1):1-7.
- Elhakim E, Mohamed O, Elazouni I. Virulence and proteolytic activity of entomopathogenic fungi against the two-spotted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae). Egypt J Biol Pest Control. 2020;30(1):1-8.
Crossref - Geoffry K, Achur RN. Screening and production of lipase from fungal organisms. Biocatal Agric Biotechnol. 2018;14:241-253.
Crossref - Roy S, Parvin R, Ghosh S, Bhattacharya S, Maity S, Banerjee D. Occurrence of a novel tannase (tan B LP) in endophytic Streptomyces sp. AL1L from the leaf of Ailanthus excelsa Roxb. 3 Biotech. 2018;8(1):33.
Crossref - Mahapatra S, Banerjee D. Extracellular tannase production by endophytic Hyalopus sp. The Journal of General and Applied Microbiology. 2009;55(3):255-259.
Crossref - Umar A, Ahmed S. Optimization, purification and characterization of laccase from Ganoderma leucocontextum along with its phylogenetic relationship. Sci Rep. 2022;12(1):2416.
Crossref - Malubag AG, Parayao AM, Waing KGD. Identification of endophytic fungi associated with banana (Musa paradisica L.) and evaluation of its enzymatic abilities. Int J Agric Technol. 2021;17(3):941-958.
- Chmelova D, Legerska B, Kunstova J, Ondrejovie M, Miertus S. The production of laccases by white-rot fungi under solid-state fermentation conditions. World J Microbiol Biotechnol. 2022;38(2):21.
Crossref - Mustafa A, Karmali A, Abdelmoez W. A sensitive microplate assay for lipase activity measurement using olive oil emulsion substrate: modification of the copper soap colorimetric method. J Oleo Sci. 2016;65(9):775-784.
Crossref - Mefteh BF, Frikha F, Daoud A, et al. Response surface methodology optimization of an acidic protease produced by Penicillium bilaiae isolate TDPEF30, a newly recovered endophytic fungus from healthy roots of date palm trees (Phoenix dactylifera L.). Microorganisms. 2019;7(3):74.
Crossref - Cavalcanti RMF, de Oliveira Ornela PH, Jorge JA, Guimaraes LHS. Screening, selection and optimization of the culture conditions for tannase production by endophytic fungi isolated from Caatinga. J Appl Biol Biotechnol. 2017;5(1):001-009.
- Chanyal S, Agrawal PK. Preliminary screening for laccase-producing endophytic fungi from Cupressus torulosa D. Don. Int J Sci Eng Manage. 2016;1:85-89.
- El-Gendi H, Saleh AK, Badierah R, Redwan EM, El-Maradny YA, El-Fakharany EM. A comprehensive insight into fungal enzymes: Structure, classification, and their role in mankind’s challenges. J Fungi. 2022;8(1):23.
Crossref - Meenashree B, Kathiravan G, Kathireshan AK. Enumeration of endophytic fungi from Eichhornia crassipes root and preliminary screening for tannase enzyme production. Asian J Microbiol Biotech Env Sc. 2020;22(2):329-333.
- D’Souza PS, Mansy TK, Preethi TC, Gunashree BS. Tamarindus indica seed induced tannase production from Aspergillus niger. Biomedicine. 2022;42(4):752-756.
Crossref - Pakaweerachat P, Chysirichote T. Valorization of tannin-rich triphala waste for simultaneous tannase and gallic acid production under solid-state fermentation by Aspergillus niger. Chem Eng Commun. 2022;210(9):1420-1433.
Crossref - Liu TPSL, Brandao Costa RMP, de Vasconcelos Freitas DJ, et al. Tannase from Aspergillus melleus improves the antioxidant activity of green tea: purification and biochemical characterisation. Int J Food Sci Technol. 2017;52(3):652-661.
Crossref - de Las Rivas B, Rodriguez H, Anguita J, Munoz R. Bacterial tannases: classification and biochemical properties. Appl Microbiol Biotechnol. 2019;103:603-623.
Crossref - Costa AM, Ribeiro WX, Kato E, Monteiro ARG, Peralta RM. Production of tannase by Aspergillus tamarii in submerged cultures. Braz Arch Biol Technol. 2008;51(2):399-404.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.