ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus (S.aureus) and coagulase negative Staphylococci (CONS) are the commonest pathogens that lead to severe bacterial infections. It is a bacterium with consistent resistance development against commonly used antibiotics, with emergence of Methicillin resistant staphylococcus aureus (MRSA) causing several infections in patients following hospitalization. Glycopeptides like vancomycin is used as primary drug for treating infectious diseases caused by MRSA. Due to indiscriminate use of vancomycin to treat MRSA, several strains with variable susceptibility to the same have emerged. Evaluation of Vancomycin Minimum Inhibitory Concentration (MIC) in the MRSA isolates obtained from clinical samples received in the diagnostic microbiology laboratory. About 120 Staphylococci obtained from different clinical samples in the diagnostic Microbiology laboratory, at tertiary health care center, South India, were included in the study. The isolates were identified and susceptibility to the relevant antibiotics was done by Vitek 2 an automated system. Vancomycin MIC was detected by Vitek 2 and E-test strip technique. Out of 120 Staphylococcal strains, 79(65.8%) S. aureus and 41(34.1%) CONS were isolated. Methicillin resistance was observed in 38 (48.1%) strains of S. aureus. Almost all 38 MRSA isolates were vancomycin sensitive with MIC range of 0.5 – 2µg/ml. Maximum isolates had MIC of 1 µg/ml i.e. 65.78% and 71% by E-Test and Vitek 2 respectively. The reported increased MIC of Vancomycin, though within the susceptible range, might experience poor clinical outcomes. Emergence and spread of resistance to glycopeptides like vancomycin needs to be kept in check by rapidly detecting the strains for resistance and strictly obeying the infection control practices.
Methicillin resistant staphylococcus aureus (MRSA), Vancomycin, Minimum Inhibitory Concentration (MIC)
Staphylococci are the normally colonizers of various skin and mucous surfaces in humans. They play a major role in several infectious diseases ranging from mild skin and soft tissue infections to serious ailments like bacteremia, endocarditis and many more.1 S. aureus is the commonest bacterial agent causing illnesses which may be acquired in the community or after hospitalization which readily acquire resistance to multiple antibiotics.2
In the year 1960’s, soon after the introduction of methicillin in clinical world mainly for treating penicillinase producing S.aureus infections, methicillin resistant Staphylococcus aureus (MRSA) emerged and spread worldwide. S. aureus which is resistant to antibiotics, especially to methicillin, are equally adapted to hospitals and outer environments evolving as a global pathogen of public health concern.3 Vancomycin is currently the drug to be chosen for treating many MRSA infections.4 However, increasing minimum inhibitory concentration (MIC) of Vancomycin, though falls in the range of susceptibility, is reported worldwide.5
Hence, the present study was carried to evaluate the vancomycin MIC values of strains of MRSA obtained from clinical samples.
This study was carried out in the Diagnostic Microbiology laboratory ,at a tertiary health care center, south India for one year duration.
Staphylococci isolated from several clinical samples like pus, blood, urine, endotracheal aspirates sent for culture and sensitivity examination for one year duration were included for the study. Samples were inoculated onto appropriate culture media and kept for overnight incubation in aerobic condition at 37°C. Blood samples received for bacterial culture were incubated in BacTec automated blood culture system and sub-cultured on the plating media if positive. Cultural characteristics suggestive of Staphylococci were analyzed on different media by microscopic examination, catalase test and later identification of Staphylococcus aureus and Antibiotic susceptibility testing for relevant antibiotics was done as per to standard protocols6 through Vitek 2 automated culture Identification and antibiotic sensitivity system by using Gram positive cartridges.
Antibiotics were tested for susceptibility by following Clinical and Laboratory Standards Institute (CLSI) guidelines7, which include Ampicillin, Erythromycin, Clindamycin, Cefoxitin, Tetracycline, Cotrimoxazole, Ciprofloxacin, Levofloxacin, Linezolid, Vancomycin, Teicoplanin, and Gentamycin.7
Methicillin-resistance was noted for the above isolates by Vitek-2 system by cefoxitin screen.
Vancomycin MIC was detected by an automated system- Vitek 2 compact, also by E-test strip method using Vancomycin EZY MICTMStrip (E test strips), procured from Himedia.
Susceptibility to the above mentioned antibiotics was interpreted by following CLSI criteria. For the quality check, ATCC 25923 S.aureus strain was used.
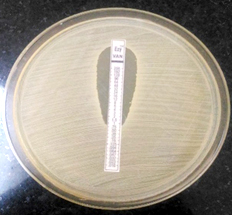
E-strip test was performed on Mueller-Hinton agar plate. Inoculum was suspended in Trypton Soy broth and incubated for 2 hours. Lawn culture of the bacterium was made on the entire plate by matching the inoculum turbidity to 0.5 McFarland standard and aseptically E-strip of Vancomycin was applied onto inoculated surface of the agar plate gently according to the manufacturer’s protocol. Following overnight incubation, a tear drop-shaped zone of inhibition intersecting the graded concentration of the antibiotic was observed. Vancomycin MIC was read at this point and results were interpreted. 7,8
In the present study, about 120 staphylococcal isolates obtained from clinical samples received in the diagnostic microbiology laboratory at a tertiary health care center, South India were studied and analyzed.
Out of 120 isolates, 81(67.5%) were isolated from pus samples, 16(13.3%) from Blood samples, 20(16.6%) from Urine samples and 3(2.5%) from ET samples.
Out of the 120 staphylococci, 79(65.8%) S. aureus and 41(34.1%) CONS were isolated.
On further analysis, 38 (48.1%) strains of S. aureus found to be methicillin resistant.
Sensitivity to relevant antibiotics for the above isolates done through Vitek 2 automated system. Of the 38 MRSA isolates, all the 38(100%) isolates were sensitive to Vancomycin and Linezolid, 35(92%) to Teicoplanin, 24(63.15) to Tetracycline, 23(60.5) to Levofloxacin, 22(57.89) to Gentamycin, 20(52.6) to Clindamycin, 15(39.47) to Cotrimoxazole, 13(34.2) to Erythromycin, 12(31.5) to Ciprofloxacin. All the strains of MRSA found resistant to Ampicillin and Cefoxitin in Vitek 2 automated system.
In the recent years, Staphylococcus aureus, more so MRSA a gram positive pathogen is becoming a global challenge causing wide range of infections both at the community and hospital setup.9 The overall prevalence of methicillin-resistant Staphylococcus aureus (MRSA) is alarmingly high and making the vancomycin (VAN) a mainstay of therapy for life threatening MRSA infections.10 Indiscriminate usage of vancomycin is implicated in the emergence and spread of resistance to the same among the MRSA isolates is a great worry in the health care settings. Minimum Inhibitory concentration (MIC) is the right indicator while selecting a proper antibiotic for treating serious infections.5
Over the last several years, many studies that have revealed an association of failure of vancomycin therapy even though MIC values were 1.5 or 2µg/ml which lie within the range of Clinical and Laboratory Standards Institute’s (CLSI’s) and the FDA’s acceptable values (32 µg/ml).10
In the present study, about 79(65.8%) S. aureus strains were obtained from clinical samples like pus, blood, urine, endotracheal aspirates.
In the current study, 38 isolates (48%) were methicillin resistant (MRSA) which is almost marginally same as mentioned by Reema et al. (46%)9 and Mita D et al. (47.5%)1. Raghabendra adhikari et al. reported 35.5%3 Nasiru abdullahi et al. 26.9%11, Radhika rana et al. 52%12, Kumari N et al. 26%13, Kala Yadav et al.1466.84% MRSA isolates in their study .
In the antibiotic susceptibility report of Vitek 2 automated system for the MRSA strains in the present study, maximum sensitivity was seen with Linezolid and Vancomycin (100%), followed by Teicoplanin (92%). This is similar to studies conducted by Mita et al.1, Reema et al.9 and Kumari et al.13 So our study highlights that same drugs appears to be the most effective therapeutic options for treating infections caused by MRSA strains.
In the present study, only 63.15% strains of MRSA were sensitive to Tetracycline, 60.5% to Levofloxacin, 57.89 % to gentamycin, 52.6% to Clindamycin. Only 39.47% were sensitive to Cotrimoxazole, 34.2% to Erythromycin, 31.5% to Ciprofloxacin. Variable rates of susceptibility to the same antibiotics were reported by Kumari et al.13 and Trivedi et al.15 This is probably due to the random usage of these drugs for empirical therapy which are cheaper and easily available to general population as over the counter drugs even without clinician prescription.
All the strains of MRSA showed resistance to ampicillin and Cefoxitin.
In the current study vancomycin MICs of MRSA isolates were measured by E- strip Test & Vitek 2 method (MIC measured in µg/ml). All the strains of MRSA found susceptible to vancomycin (MIC£ 2µg/ ml) by both the methods.
In our study MIC of Vancomycin by E- strip test was fall in the range of 0.5 –2µg/ml which is similar to and very well correlated to studies conducted by Ranjan et al.16 and Mouton et al.17
Robin et al., reported Vancomycin MIC values of 8-16mg/l by MRSA strains in their trial which were found to be resistant to Vancomycin.18
Vancomycin MIC detected by two different methods vitek 2 and E-Test method in the present study are almost similar with slight variation. About 18.4% isolates showed MIC of 0.5µg/ml, 65.78% with 1µg/ml and 15.78% strains showed MIC value of 2µg/ml by E- strip Test. Whereas with Vitek 2 method, 15.7% revealed MIC value of 0.5µg/ml, 71% strains with 1µg/ml & 13.15% showing 2µg/ml of MIC.
Diaz et al.4 also reported no significant differences between E-Test method & BMD for vancomycin MIC detection for MRSA isolates.
In a study done by Himani et al.19 E-test corelated better with BMD method than Vitek 2 preferring E-Test method for determining vancomycin MICs than Vitek 2.
Table (1):
Showing the Antibiotic susceptibility of the staphylococci
Antibiotics |
MRSA (n=38) |
MSSA (%) (n=41) |
|---|---|---|
Ampicillin |
00(00) |
18(43.9) |
Erythromycin |
13(34.2) |
27(65.8) |
Clindamycin |
20(52.6) |
31(75.6) |
Cefoxitin |
00(00) |
41(100) |
Tetracycline |
24(63.15) |
32(78) |
Cotrimoxazole |
15(39.47) |
30(73.17) |
Ciprofloxacin |
12(31.5) |
30(73.17) |
Levofloxacin |
23(60.5) |
32(78) |
Linezolid |
38(100) |
41(100) |
Vancomycin |
38(100) |
41(100) |
Teicoplanin |
35(92) |
41(100) |
Gentamicin |
22(57.89) |
31(75.6) |
Table (2):
MIC of MRSA isolates to Vancomycin
MIC value of Vancomycin |
Vitek 2 (n=38) |
E-strip (n=38) |
|---|---|---|
0.5 |
06(15.7%) |
07(18.4%) |
1 |
27(71%) |
25(65.78%) |
2 |
05(13.15%) |
06(15.78%) |
4 |
0 |
0 |
Out of 38 MRSA isolates, 07(18.4%) showed MIC of 0.5μg/ml, 25(65.78%) isolates had MIC of 1 μg/ml and 06(15.78%) isolates had MIC of 2 μg/ml by E- strip Test. By Vitek 2 method, 6 isolates (15.7%) had 0.5μg/ml of MIC & 27 isolates (71%) had 1 μg/ml of MIC, 05(13.15%) isolates had 2μg/ml of MIC.
The reported increased minimum inhibitory concentration (MIC) of Vancomycin, though falls within the values indicating susceptibility, might experience poor clinical outcomes. Upon reporting of higher MIC values, additional doses of the antibiotics over the indicated one to obtain the desired drug concentrations were administered with reduced success rates, more days of hospital stay and higher rates of mortality. Prolonged exposure of bacteria to sub inhibitory vancomycin levels is believed to be a crucial factor for rising levels of vancomycin MIC. The emergence and spread of vancomycin resistant strains is a menace to the demanding modality of MRSA treatment and seting up a worrying situation to the treating clinicians.
Ongoing efforts are necessary to combat emergence and dissemination of resistance to glycopeptide agents by adhering strictly to the infection control strategies and timely recognition of the resistant strains in the health care set up.
Acknowledgements
None.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
All authors have made substantial contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
The study was undertaken after obtaining approval from the Institutional Research and Ethics Committee.
- J.V. Sathish and Mita D. Wadekar, Prevalence of MRSA in Clinical Samples and their Antibiotic Sensitivity Pattern, Int. J. Curr. Microbiol. App. Sci., 2017; 6(7): 2338-2343.
Crossref - Jayshree, Vinod Kumar Singh, Amit Kumar and Sharad Kumar Yadav- Prevalence of methicillin resistant Staphylococcus aureus at tertiary-care hospital (MRSA) of Mathura , India – Progressive Research – An International Journal, 2016; 11(Special-VIII): 5495-5498.
- Raghabendra Adhikari, Narayan Dutt Pant, Sanjeev Neupane, Mukesh Neupane, Roshan Bhattarai, Sabita Bhatt et al., Detection of Methicillin Resistant Staphylococcus aureus and Determination of Minimum Inhibitory Concentration of Vancomycin for Staphylococcus aureus Isolated from Pus/Wound Swab Samples of the Patients Attending a Tertiary Care Hospital in Kathmandu, Nepal, Hindawi Canadian Journal of Infectious Diseases and Medical Microbiology, 2017: 1-7.
- R. Diaz, V. Afreixo, E. Ramalheira, C. Rodrigues, B. Gago, Evaluation of vancomycin creep in Methicillin- Resistant Staphylococcus aureus infection-a systematic review & meta-analysis. Clinical Microbiology and Infection, 2018; 24: 97e104.
Crossref - Bansidhar Tarai, Poonam Das, Dilip Kumar, Recurrent Challenges for Clinicians: Emergence of Methicillin resistant Staphylococcus aureus, Vancomycin Resistance, and Current Treatment Options. J. of Laboratory Physicians, 2013; 5(2).
Crossref - Collee JG, Fraser AG, Marmion BP, Simmons A. 2007 Mackie and McCartney practical medical Microbiology. 245-269, 14th edition. Churchill Livingstone
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 26thed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
- Forbes BA, Sahm DF, Weissfeld AS. Bailey and Scott”s Diagnostic Microbiology. 2007; 187-214, 12th ed. Mosby Elsevier.
- Hannath Ayesha Reema, Saldanha Dominic RM, Prevalence and antimicrobial susceptibility pattern of clinical isolates of Methicillin-resistant Staphylococcus aureus in a tertiary care hospital in Mangalore, Journal of International Medicine and Dentistry, 2016; 3(3): 134-139.
Crossref - Michael J. Rybak, Celine Vidaillac, Helio S. Sader, Paul R. Rhomberg, Hossein Salimnia, Lawrence E. Briski, Audrey Wanger, Ronald N. Jonesc. Evaluation of Vancomycin Susceptibility Testing for Methicillin-Resistant Staphylococcus aureus: Comparison of E-test and Three Automated Testing methods. Journal of Clinical Microbiology, 2013; 51(7): (2077–2081).
Crossref - Nasiru Abdullahi, Kenneth Chukwuemeka Iregbu, Methicillin Resistant Staphylococcus aureus in a Central Nigeria Tertiary Hospital, Annals of Tropical Pathology ¦ January – June 2018; 9(1).
- Radhika Rana-Khara, Sucheta J. Lakhani, Sangita Vasava, Krunal Shah, Dipak Panjwani, Methicillin Resistant Staphylococcus aureus (MRSA) and Vancomycin Resistant Staphylococcus aureus (VRSA) from a rural based tertiary care and teaching hospital in Vadodara district, Gujarat. IAIM, 2016; 3(7): 187-195.
- Kumari N, Mohapatra TM, Singh Y, Prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) in a Tertiary-Care Hospital in Eastern Nepal. J. Nepal Med. Assoc., 2008; 47(170):53-6.
Crossref - Kala Yadhav M.L., Gayathri J. Panicker, Prevalence And Antibiogram Of Methicillin Resistant Staphylococcus Aureus Isolates At A Tertiary Care Hospital In Bangalore, South India, Int. J. Cur. Res. Rev., 2014; 6(17).
- Minal B Trivedi, Mahendra Vegad, Sumeeta Soni, Prevalence of methicillin-resistant Staphy-lococcus aureus in various clinical samples in a tertiary-care hospital , International Journal of Medical Science and Public Health, 2015; 4(12).
- KP Ranjan, DR Arora, Neelima Ranjan, An Approach to Linezolid and Vancomycin against Methicillin Resistant Staphylococcus Aureus”, Webmed Central MICROBIOLOGY, 2010; 1(9): WMC00590.
- J.W. Mouton and A. R. jansz, The Duel study: a multi-center in vitro evaluation of Linezolid compared with other Antibiotics in the Netherlands. Journal European Society of Clinical Microbiology and Infectious Disease 2001, 486-491.
Crossref - Robin A. Howe, Alastair Monk , Mandy Wootton , Timothy R. Walsh and Mark C. Enright at Southmead Hospital, Bristol, United Kingdom, Vancomycin susceptibility with in Methicillin- resistant Staphylococcus aureus Lineages, Emerging infectious Disease, 2004; 10: 855-857.
Crossref - Himani, Charu agrawa, Molly Madan, Anita Pandey, Bhaskar ThakuriaMethicillin Resistant Staphylococcus aureus: Inconsistencies in Vancomycin Susceptibility Testing Methods, Limitations and Advantages of each Method. Journal of Clinical and Diagnostic Research, 2015; 9(10): DC01-DC04.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.