ISSN: 0973-7510
E-ISSN: 2581-690X
A total of 13 Actinomycete strains were isolated from 70 soil samples collected from five locations across the Jeddah Province, while the other two locations located in Baljurashi province of Saudi Arabia. All 13 isolates were purified and subjected to enzymatic screening and antibacterial assays. The results indicated that two of these isolates (AC45 and AC69) produced both enzymes and exerted some antibacterial activity. Isolate AC45 produced more amylase and polygalacturonase (697.8 and 1498.59 units/ml, respectively) than isolate AC69; however, AC69 secreted more lipase than AC45 (6957 and 22127 unit/ml, respectively). Furthermore, both AC45 and AC69 exhibited good antibacterial activity against Staphylococcus aureus, Staphylococcus epidermidis, and Bacillus subtilis. The two isolates were identified using their 16S rRNA sequences, and the results suggest that isolate AC45 shares 99.71% similarity with Streptomyces lavenduligriseus and isolate AC69 shares 99% similarity with Streptomyces sp.
Antibacterial, Actinomycetes, Amylase, Polygalacturonase, Lipase
Actinomycetes are filamentous gram-positive bacteria1, which produce hyphae2 and are characterized by the production of conidia3. They also have a high G-C content, at over 55% of their DNA3,4, are unicellular2 and non-motile1. The Actinobacteria phylum is one of the largest taxonomic units within the bacterial domain5,6 with its constituents widely distributed across different environments7-9, including some extreme habitats10. Despite this, soil remains the most significant habitat for Actinomycetes species2.
Actinobacteria have the ability to produce various natural bioactive products10, which are both economically and biologically valuable, including enzymes, antibiotics, antitumor, and immune regulatory agents11-13. In fact, two-thirds of the world’s most common antibiotics are synthesized by Actinobacteria14, with most of these products produced by the members of the Streptomyces genus4.
Various studies have reported that some genera of the Actinomycetes produce a diverse range of enzymes that can be applied in biotechnology driven industries and bio microbial applications15. In 1940, the first Actinomycete derived antibiotic (Actinomycin) with bacteriostatic and bactericidal properties was purified from Streptomyces antibioticus (formerly known as Actinomyces antibioticus)16-20.
With the spread of resistance of certain microbes to antibiotics, the need to discover new antibiotics is increasing. Actinomycete is one of the possible ways to increase the chance of finding new and interesting antibiotics due to their diversity.
The searching for a new site, such as the deserts to take soil samples and isolate some of the Actinomycetes for the studying of secondary metabolites, which can be detected for some of the reliable materials in the field of resistance to pathogenic bacteria rather than the most common types of Streptomyces found in soil samples around the world21 22.
Here we isolated a series of Actinomycete strains from the desert soil from five locations in Saudi Arabia. These isolates were evaluated for their enzyme (a-amylase, polygalacturonase, and lipase) production and antimicrobial activity against several known pathogens. Identification of the selected isolates was performed using their 16S rRNA sequences.
Sample collection
A total of 70 soil samples were collected from five different locations across Saudi Arabia including three sites in the Jeddah Province and two sites in the Baljurashi province (Table 1). Samples were collected at a depth of 15 cm using a shovel and stored in sterile polyethylene bags. The samples were then transported to the laboratory for further isolation, where remained until dryness in room temperate for three days.
Table (1):
Locations of the samples.
| Location | GPS reading | No. of samples | |
|---|---|---|---|
| Jeddah | 1 | N 21º50’22.5 E 39º08’12.7 | 20 |
| 2 | N 21º54’15.3 E 39º21’04.9 | 13 | |
| 3 | N 21º53’36.1 E 39º21’00.9 | 12 | |
| Baljurashi | 4 | N 19º53’22.7 E 41º35’29.5 | 14 |
| 5 | N 19º53’32.0 E 41º35’31.5 | 11 | |
Isolation of Actinomycetes
Actinomycetes were isolated using the soil dilution plate technique23 and Starch-Casein agar (SCA) plates24. Briefly, one gram of soil sample was suspended in 10 ml of distilled water and then diluted up to 10-5 “ dilutions4. Then, 100 µl of each dilution was spread on the SCA medium and incubated at 28 °C for 7–15 days.
Screening for enzymes production
Three types of enzymes were evaluated in this study, the assay for each is described below.
α-Amylase
a-Amylase activity was determined by measuring the amount of maltose released from a starch substrate following addition to the growth media25. The absorbance was recorded at 560 nm using a spectrophotometer (Jenway 6305 UV/Visible Spectrophotometer) and one unit of enzyme activity was defined as the amount of enzyme producing 1 µmol of reducing sugar as maltose per hour under standard assay conditions.
Polygalacturonase
Polygalacturonase activity was assayed using polygalacturonic acid as a substrate26. The absorbance was measured at 560 nm using a spectrophotometer (Jenway 6305 UV/Visible Spectrophotometer) and one unit of enzyme activity was defined as the amount of enzyme that liberated 1 ìmol of galacturonic acid per minute under standard assay conditions.
Lipase
Lipase activity was assayed using the Winkler and Stuckmann spectrophotometric assay27. The absorbance of the released p-nitrophenol was recorded at 410 nm using a spectrophotometer (Jenway 6305 UV/Visible Spectrophotometer).
Extraction of the secondary metabolites
Two isolates were selected to fermented in liquid medium (starch-casein) in shaking incubator on 180rpm at 30°C for 7 days. The mycelia were collected and added to methanol in ratio of 1:1 (v/v), and shaking it overnight, after that, the methanol extract was evaporated in a rotary evaporated until dryness. The culture filtrate (200 ml) was extracted by adding ethyl acetate as a solvent where were added to the in the ratio of 1:1 (v/v). followed by shaken vigorously for 10 minutes. The ethyl acetate phase was separated from the aqueous phase using separating funnel then evaporated in a rotary evaporated until dryness.
Screening for Antimicrobial Activities
The antimicrobial activity of the selected Actinomycetes isolates was screened using two pathogenic bacteria (Staphylococcus aureus ATCC 6538 and Staphylococcus epidermidis ATCC 12228) and one Bacillus subtilis isolated obtained from King Abdul Aziz Hospital in Jeddah, Saudi Arabia. Antimicrobial efficacy was determined using the agar-well diffusion method, where 6 mm diameter wells were created in the agar media and then filled with the antimicrobial extract28.
Mueller Hinton agar plates were prepared and inoculated with the bacteria of interest and allowed to grow to a lawn culture before the wells were filled with the extract of interest. The plates were then cooled at 4 °C for 30 seconds and then incubated at 37 °C for 24 h. Microbial inhibition was observed as zones of clearance which were then measured and recorded.
Isolate identification
The DNA from each isolate was extracted and sent to macrogene, in Seoul, Republic of Korea where it underwent 16s rRNA sequencing. These sequences were then compared to the 16S rRNA sequences in the NCBI GenBank database using the basic alignment search tool (BLAST)29.
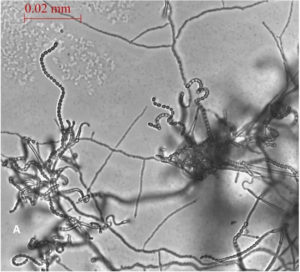
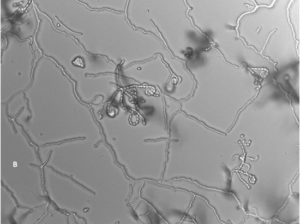
A total of 13 Actinomycetes were isolated from 70 soil samples collected from five different locations across Saudi Arabia. Among the isolates, two strains (AC 45 and AC 69) demonstrated both enzyme production and antimicrobial activity against at least one of the bacterial pathogens used in the preliminary screening. BLAST analysis of the 16S rRNA gene sequences from these soil Actinomycetes isolates revealed that AC45 shared 99.71% sequence similarity with Streptomyces lavenduligriseus and AC69 shared 99% sequence similarity with Streptomyces sp. G49 These results are summarized in Table 2 and the morphological structure for each isolate is described in Figure 1.
Table (2):
List of BLAST results for the 16S sequence.
Isolate no. |
Description |
Accession |
|---|---|---|
AC 45 |
Streptomyces lavenduligriseus strain NRRL ISP-5487 contig85.1 |
NZ_JOBD01000085.1 |
AC 69 |
Streptomyces sp. G49 |
KT741025.1 |
Fig. 1. Spore chain arrangement observed at 1000X. A) Isolate AC 45- Streptomyces lavenduligriseus, B) Isolate number AC 69 – Streptomyces sp.
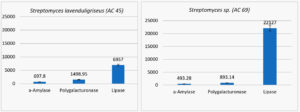
The two isolates were screened for production of a-amylase, polygalacturonase, and lipase, where the estimation of enzymes productivity are recorded as (unit/ml) Figure 2.
Fig. 2. The estimation of enzymes productivity (unit/ml) for two isolate of streptomyces (Streptomyces lavenduligriseus and Streptomyces sp.)
The S. lavenduligriseus isolate was shown to produce more amylase and polygalacturonase than the Streptomyces sp isolate but the Streptomyces sp isolate secreted more lipase than the S. lavenduligriseus isolate.
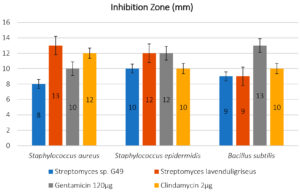
The antimicrobial activities of the selected isolates were tested against several pathogenic strains, and antagonism was measured by determining the size of the zone of inhibition (mm) for each (Figures 3 and 4).
Fig. 3. A: Streptomyces lavenduligriseus (AC 45) was recorded a moderate effect against Staphylococcus epidermidis while Streptomyces sp.(AC 69) showed a good effect. B: isolate of Streptomyces sp. (AC 69) showed a strong effect against Staphylococcus aureus while isolate S. lavenduligriseus (AC 45) showed a moderate effect. C: the same result in B but in the presence of Bacillus subtilis.
The results, which illustrated in Figure 4 that showing the isolate of Actinomycetes Streptomyces sp. G49 which was recorded the best result for inhibition pathogenic bacterial growth in all treatments to S. aureas, S. epidermidis and B. subtilis, (8, 10, 9 mm) respectively, comparing with the results for using standard antibiotics, Gentamicin 120ug (10,12, 13mm), and Clindamycin 2ug (12, 10 and 10mm) respectively.
Fig. 4. Antagonistic activity of Actinomycetes isolates by mm against pathogens (Staphylococcus aureus ATCC 6538; Staphylococcus epidermidis ATCC 12228; Bacillus subtilis) with 125 μl of extract comparing with two stander antibiotic Gentamicin 120μg and Clindamycin 2 μg.
On the other hand, the isolate of S. lavenduligriseus outperformed on the both antibiotics to control of Bacillus subtilis, which recoded 9 mm (inhibition zone size) compared by 13 and 10mm for both Gentamicin 120ug and Clindamycin 2ug, respectively.
This study successfully isolated two antibacterial producing Actinomycetes from Saudi desert soils. Among the 13 selected isolates, only two demonstrated the ability to produce antibacterial agents with activity against the tested microbes, and both AC45 (Streptomyces lavenduligriseus) and AC69 (Streptomyces sp.) showed good antibacterial activity against Bacillus subtilis, Staphylococcus aureus, and Staphylococcus epidermidis. These results were similar to the study conducted by Ganesan et al4, who screened 106 isolates from Nilgiris district soil in the Western Ghats of Tamil Nadu, India, and cross-streaked against various microbial pathogens, and found that 44 isolates exhibited good antimicrobial activity against different pathogenic microbes4. In addition, Mohamed et al30 isolated 32 Actinomycetes from Algerian desert soil, three of which showed antibacterial activity against B. cereus, S. aureus, and S. epidermidis.30
Both isolates were also evaluated for the production of a-amylase and both isolates demonstrated good a-amylase activity, which is in agreement with the study conducted by Yassien and Asfour31 who demonstrated the ability of five Streptomyces isolates to produce a-amylase31. Our isolates also produce polygalacturonase and lipase, which is in agreement with the study conducted by Salehghamari et al32 on a Streptomyces coelicoflavus isolate from the Meyghan Salt Lake of Arak located in the Markazi Province of Iran, which produced polygalacturonase, and Cardenas et al33, who identified three novel lipases produced by Actinomycetes strains32,33. These results are also in agreement with Swarna and Gnanadoss34 who identified five lipases producing from Actinomycetes34.
In future studies, both enzymes and antimicrobials component will be extracted and purified with the aim of determining the chemical composition, to try to use it as anti-pathogenic materials as one of the attempts to find new types of natural antibiotics produced from desert Actinomycetes.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
- Sharma M, Dangi P, Choudhary M. Actinomycetes: source, identification, and their applications. Int J Curr Microbiol App Sci. 2014;3(2):801-32.
Crossref - Anandan R, Dharumadurai D, Manogaran GP. An introduction to actinobacteria. Actinobacteria-Basics and Biotechnological Applications: IntechOpen. 2016.
- Chaudhary HS, Yadav J, Shrivastava AR, Singh S, Singh AK, Gopalan N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India). Journal of advanced pharmaceutical technology & research. 2013;4(2):118.
Crossref - Ganesan P, Reegan AD, David RHA, et al. Antimicrobial activity of some actinomycetes from Western Ghats of Tamil Nadu, India. Alexandria journal of medicine. 2017;53(2):101-10.
Crossref - Barka EA, Vatsa P, Sanchez L, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016;80(1):1-43.
Crossref - Ludwig W, Euzéby J, Schumann P, et al. Road map of the phylum Actinobacteria. Bergey’s manual® of systematic bacteriology: Springer, 2012:1-28.
Crossref - Kumar PS, Duraipandiyan V, Ignacimuthu S. Isolation, screening and partial purification of antimicrobial antibiotics from soil Streptomyces sp. SCA 7. The Kaohsiung journal of medical sciences. 2014;30(9):435-46.
Crossref - Ahmad MS, El-Gendy AO, Ahmed RR, Hassan HM, El-Kabbany HM, Merdash AG. Exploring the Antimicrobial and Antitumor Potentials of Streptomyces sp. AGM12-1 Isolated from Egyptian Soil. Frontiers in Microbiology. 2017;8(438).
Crossref - Goodfellow M, Williams S. Ecology of actinomycetes. Annual review of microbiology. 1983;37(1):189-216.
Crossref - Qin S, Li W-J, Klenk H-P, Hozzein WN, Ahmed I. Actinobacteria in Special and Extreme Habitats: Diversity, Function Roles and Environmental Adaptations. Frontiers in microbiology. 2019;10:944.
Crossref - Mukhtar S, Zaheer A, Aiysha D, Malik KA, Mehnaz S. Actinomycetes: a source of industrially important enzymes. J Proteomics Bioinform. 2017;10:12.
Crossref - Prakash D, Nawani N, Prakash M, et al. Actinomycetes: a repertory of green catalysts with a potential revenue resource. BioMed research international. 2013;2013.
Crossref - Kamjam M, Sivalingam P, Deng Z, Hong K. Deep Sea Actinomycetes and Their Secondary Metabolites. Frontiers in Microbiology. 2017;8(760).
Crossref - Jakubiec-Krzesniak K, Rajnisz-Mateusiak A, Guspiel A, Ziemska J, Solecka J. Secondary metabolites of actinomycetes and their antibacterial, antifungal and antiviral properties. Pol. J. Microbiol. 2018;67:259-72.
Crossref - Harir M, Bendif H, Bellahcene M, Fortas Z, Pogni R. Streptomyces Secondary Metabolites. Basic Biology and Applications of Actinobacteria. 2018:99.
Crossref - Law JW-F, Pusparajah P, Ab Mutalib N-S, Wong SH, Goh B-H, Lee L-H. A Review on Mangrove Actinobacterial Diversity: The Roles of Streptomyces and Novel Species Discovery. Progress In Microbes & Molecular Biology. 2019;1(1).
Crossref - Ser H-L, Tan LT-H, Law JW-F, et al. Focused Review: Cytotoxic and Antioxidant Potentials of Mangrove-Derived Streptomyces. Frontiers in Microbiology. 2017;8(2065).
Crossref - Sakula A. Selman Waksman (1888-1973), discoverer of streptomycin: a centenary review. British journal of diseases of the chest. 1988;82:23-31.
Crossref - Waksman SA, Woodruff HB. The soil as a source of microorganisms antagonistic to disease-producing bacteria. Journal of bacteriology. 1940;40(4):581.
Crossref - Waksman SA, Woodruff HB. Actinomyces antibioticus, a new soil organism antagonistic to pathogenic and non-pathogenic bacteria. Journal of bacteriology. 1941;42(2):231.
Crossref - Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Current opinion in pharmacology. 2008;8(5):557-63.
Crossref - Tiwari K, Gupta RK. Diversity and isolation of rare actinomycetes: an overview. Critical Reviews in Microbiology. 2013;39(3):256-94.
Crossref - Johnson CF, Curl EA, Bond JH, Fribourg HA. Methods for studying soil microflora-plant disease relationships. Burgess Publishing, Minneapolis 1959.
- Küster E, Williams S. Selection of media for isolation of streptomycetes. Nature. 1964;202(4935):928.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry. 1959;31(3):426-28.
Crossref - Pathak N, Sanwal G. Multiple forms of polygalacturonase from banana fruits. Phytochemistry. 1998;48(2):249-55.
Crossref - Winkler UK, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. Journal of bacteriology. 1979;138(3):663-70.
Crossref - Al-Dhabi NA, Esmail GA, Duraipandiyan V, Arasu MV. Chemical profiling of Streptomyces sp. Al-Dhabi-2 recovered from an extreme environment in Saudi Arabia as a novel drug source for medical and industrial applications. Saudi journal of biological sciences. 2019;26(4):758-66.
Crossref - Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215(3):403-10.
Crossref - Mohamed H, Miloud B, Zohra F, Garcia-Arenzana JM, Veloso A, Rodriguez-Couto S. Isolation and Characterization of Actinobacteria from Algerian Sahara Soils with Antimicrobial Activities. Int J Mol Cell Med. 2017;6(2):109-20
Crossref - Yassien M, Asfour HZ. Production of Alpha-amylase by Streptomyces species isolated from west area of Saudi Arabia. J. Pharm. Res. 2011;4(12):4393-95
- Salehghamari E, Nasrollahzadeh Z, Tahmaseb M, Amoozegar MA. Pectinase enzyme from Streptomyces coelicoflavus GIAL86 isolated from Meyghan Salt Lake, Arak, Iran. International Journal of Aquatic Biology. 2019;7(2):106-11
- Cardenas F, Alvarez E, De Castro-Alvarez M, Sánchez-Montero J, Elson S, Sinisterra J. Three new lipases from actinomycetes and their use in organic reactions. Biocatalysis and Biotransformation. 2001;19(4):315-29.
Crossref - Swarna D, Gnanadoss JJ. Screening and Molecular Characterization of Actinomycetes from Mangrove Soil Producing Industrially Important Enzymes. Journal of Scientific Research. 2020;64(2).
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.