ISSN: 0973-7510
E-ISSN: 2581-690X
Lignocellulosic residues, including rice stubble, carboxymethyl cellulose (CMC), xylan, and lignin, were evaluated as substrates for cultivating lignocellulolytic fungi in submerged fermentation at 30 °C over 15 days. Rice stubble, accounting for 40-60% of rice plant biomass and containing 42.14% cellulose, 22.08% hemicellulose, and 11.98% lignin, was explored as a renewable resource for energy and biochemical production. Four fungal strains-Penicillium oxalicum (F1), Talaromyces pinophilus (F12), Penicillium griseofulvum (F22), and Trichoderma reesei (F26) were evaluated for their lignocellulolytic enzyme production potential. Enzyme assays conducted at 3-day intervals revealed maximal production of CMCase (63.42-88.26 U/mL), FPase (46.01-80.66 U/mL), xylanase (1146.10-1640.52 U/mL), lignin peroxidase (0.192-0.287 U/mL), and laccase (0.193-0.434 U/mL). ITS (internal transcribed spacer) sequencing confirmed the fungal strain identities. These findings highlight the potential of the tested fungal strains for hydrolytic stubble production and lignocellulose degradation, positioning rice straw as a cost-effective carbon source for biotechnological applications in biorefineries.
Rice Stubble, Fungal Isolates, Degradation, Enzyme Activity, Lignocellulosic Residues
The natural world is abundant with a wide variety of microorganisms, most of which provide valuable benefits to our environment. Among these microorganisms, fungi are essential in agriculture as they contribute significantly to the breakdown of organic matter and the management of waste. Researchers have consistently focused on exploring novel fungal strains due to their potential for producing lignocellulosic enzymes, which hold significant industrial importance. Lignocellulosic enzymes, produced by various filamentous fungi, comprise a complex of hydrolytic and oxidative enzymes that decompose lignocellulosic biomass, especially crop residues.1,2 These enzymes have garnered growing research interest due to their wide substrate versatility, sustainable valorisation and usefulness in industrial applications of lignocellulosic biomass.3
Agricultural, forestry, and agro-industrial waste that typically accumulates in the environment presents significant ecological challenges. Nevertheless, lignocellulosic wastes have the potential to be utilised as important resources for the efficient manufacturing of valuable products such as secondary metabolites, organic acids, biocatalysts, biofuels, and other refined chemicals. These applications are currently attracting significant attention from researchers. Since the cost of lignocellulosic feedstock is crucial in determining the economic viability of the delignification process, considerable research has focused on using low-cost substrates.
Agricultural biomass has garnered attention as a carbon source for producing cellulase enzymes through submerged fermentation using microbial species.4 Currently, lignocellulolytic enzymes, including hydrolytic and lignolytic are widely employed in the food industry for various purposes such as texturizing and flavouring during the preparation of fruits and vegetables. Additionally, these enzymes enhance the softness of clothes by causing fibers to bulge and minimize the visibility of diminishing in fabrics.5,6
Although, as per the Indian Ministry of New and Renewable Energy (MNRE), India produces 686 million tons (MT) of dry matter agricultural residue annually from different crops. Nevertheless, there remains a surplus of 234.5 million metric tons of crop residue, which contributes to the production of excessive particulate matter and air pollution.7,8 The rising demand for rice due to a growing population has resulted in the generation of substantial amounts of rice stubble. The rice plant consists of approximately 40%-60% rice stubble, a lignocellulosic biomass containing 46.55% cellulose, 28.25% hemicellulose, 11.60% lignin, 13.70% silica, 0.81% nitrogen, 0.32% phosphorus, and 2.78% potassium.9,10 The roles of cellulase, xylanase, laccase, and lignin peroxidase in the conversion of agricultural and other organic waste have been extensively studied in recent decades.
As a result, straw can be a decent feedstock for producing cellulase enzymes. Utilizing crop residue as a feedstock in microbial fermentation processes could be a more cost-effective method for cellulase enzyme production, thereby reducing overall costs. Cellulase enzymes are secreted by microorganisms, such as bacteria and fungus, during the process of breakdown or fermentation.11 Cellulases hydrolyse cellulose by breaking β-1,4-glycosidic bonds, with endoglucanases initiating cleavage in amorphous regions, exoglucanases break processing ends of chain which release cellobiose, and lastly β-glycosidases converting cellobiose into glucose molecules. Effective cellulose hydrolysis requires synergistic action among these three enzyme types for complete and efficient conversion. Hemicellulases break glycosidic bonds between carbohydrates and aid glycohydrolases by removing surface methyl and acetyl groups. They include depolymerizing enzymes (xylanases, glucanases, mannanases) that hydrolyze main chain bonds, and accessory enzymes that target ester and side chain bonds (acetyl xylan esterase, α-L-arabinofuranosidase, glucuronyl esterase, ferulic acid esterase and β-glucuronidase). Lignin, an aromatic and hydrophobic polymer having high molecular weight, resists the action of hydrolases due to its physiological stability provide by ester and ether bonds, requiring oxidative reactions for its degradation. Ligninases, including peroxidases (lignin, manganese, versatile, and bleaching) and oxidases, facilitate this process. Peroxidases, heme-containing glycoproteins, use hydrogen peroxide to oxidize phenolic and non-phenolic compounds, forming free radicals and Mn3+ ions, which degrade lignin.6
Although, various fermentation techniques, such as solid-state, submerged, and co-culture fermentation, can be employed to produce lignolytic and hydrolytic enzymes. Optimizing these processes can enhance enzyme yield and productivity without incurring extra costs. Additionally, genetic engineering can be leveraged to boost enzyme production to satisfy industrial demands. Therefore, this study aims to screen fungal species for their ability to produce lignolytic and hydrolytic enzymes through submerged fermentation using rice stubble. The objective is to identify effective fungal strains that are suitable for large-scale enzyme production.
Procurement of the materials
Fresh rice stubbles were collected from a farmer’s field in Salarpur village, Sirsa, India. It was cut into pieces of 2-3 cm using a fodder cutter and then stored in high-quality polythene bags at the room temperature.
Isolation of lignocellulolytic fungi for degradation of rice stubbles
Rice stubble were collected from the ICAR-IARI fields in New Delhi, India, after the rice harvest. These stubble, along with soil samples, were immediately placed into RMM medium12 supplemented with 1% carboxymethyl cellulose (CMC), 1% xylan, and 0.1% lignin, and incubated at 30 °C for 14 days. After the enrichment period, fungi were isolated using the serial dilution and spread plate method on RMM medium containing 1% CMC, 1% xylan, and 0.1% lignin. Fungal isolates with distinct morphological features were selected, purified, and maintained on potato dextrose agar slants.
Maintenance of the Fungal strain
The isolated colonies were subcultured three times on fresh agar plates to ensure the cultures were purified. The fungal culture showing distinct morphology was maintained as per the growth conditions, that is, incubation temperature of 28 ± 2 °C, incubation period of 7 days and was stored in refrigerator at 4 °C after sub-culturing
Morphological observations and identification of the promising lignocellulolytic fungal strains
Morphological observations were conducted on four different fungal isolates cultured on 1.5% PDA at approximately 28 ± 2 °C. The more complex conidiophores, which developed from characteristic tufted or pustulate areas of conidiation typically 3 to 5 days after inoculation, were examined microscopically. A compound microscope equipped with a Progres 2.7 camera (Jenoptik, USA) was used to analyze the color, size, shape, and septation of the conidiophores and conidia of each strain. Measurements were taken using MagVision software, with 25 recordings per replication to ensure accuracy. The fungal strains were identified using identification keys provided by Rifai13 and Bissett.14 The fungal isolates are identified as lignocellulolytic fungal strains on the basis of their enzymatic studies.
Molecular identification of the promising lignocellulolytic fungal strains
The genetic diversity of fungal strains from the genera Trichoderma, Penicillium, and Talaromyces was examined using the random amplified polymorphic DNA (RAPD) approach.15 7 days old mycelial biomass of Trichoderma, Penicillium and Talaromyces culture were transferred to Potato dextrose broth (PDB) in aseptic condition under the laminar air flow and the cultures were then incubated at a temperature of 28 °C. The fungal mycelial mat was harvested after 8-10 days and subsequently stored at -20 °C for future study.
Isolation of genomic DNA
The fungal mycelium was pulverised into a fine powder using liquid nitrogen. Four grams of powdered mycelium were mixed with C-TAB buffer and incubated at 60 °C for one hour in a water bath. Afterwards, the mixture was subjected to centrifugation at a rate of 10,000 revolutions per minute for 10 minutes at 24 °C. Following this, 500 µl of the resulting liquid above the sediment was carefully collected and transferred to a fresh tube. Subsequently, an equal quantity of Chloroform: Isoamyl alcohol (24:1) was added, and the mixture was centrifuged again at a speed of 10,000 rpm for 2 minutes at a temperature of 24 °C. Subsequently, 500 µl of the supernatant was transferred to a separate tube. Then, 0.6 times the volume of isopropanol (300 µl for 500 µl of the liquid) and 0.1 times the volume of sodium acetate (50 µl for 500 µl of the liquid) were introduced. Subsequently, the tubes were placed in a controlled condition at -20 °C. On the following day, the tubes were subjected to centrifugation at a speed of 10,000 rpm for 10 minutes at 4 °C. The liquid portion above the solid sediment at the bottom was meticulously removed without causing any disturbance to the sediment. To wash the pellet, 500 µl of 75% ethanol was added, and the solution was subjected to another round of centrifugation at 10,000 revolutions per minute for a duration of 10 minutes at a temperature of 4 °C. The pellets were dried by exposing them to air after the ethanol was removed. The DNA was dissolved by adding TE buffer, and the solution was kept at a temperature of 4 °C for an entire night before being stored at -20 °C. The DNA concentration was determined using a Nano-Drop spectrophotometer and subsequently utilised for PCR amplification.
Amplification of ITS region, sequencing, alignment and identification
The ITS region was amplified using a Thermal cycler manufactured by Biotron Healthcare (India) Pvt. Ltd. In order to enhance the signal, the amplification process utilised Internal Transcribed Spacer (ITS) primers, namely ITS-1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS-4 (5’-TCCTCCGCTTATTGATATGC-3’), as described by White et al.16 The PCR was performed under optimised conditions. The procedure began with an initial denaturation phase at a temperature of 95 °C for a duration of 5 minutes. This was followed by a series of 30 cycles consisting of denaturation on 95 °C for 30 seconds, annealing at 50 °C for 30 seconds, and extension takes place at 72 °C for period of 60 seconds. Finally, there was a completing step of elongation at 72 °C for a period of 10 minutes. MilliQ water was used as a negative control in the experiment. PCR results were visualized on a 1.2% horizontal agarose gel containing 0.002% ethidium bromide. Gel images were captured using a Gel Documentation System (AlphaImager 1220, Alpha Innotech Corporation, California) equipped with a CCD camera. ITS rRNA sequencing was outsourced to the Eze Diagnon sequencing facility in Coimbatore, India, using the same primer set. The aligned sequences were analyzed through BLAST using the EzTaxon-e server.17 A phylogenetic tree was constructed with MEGA 6.0 software employing neighbor-joining, maximum parsimony, and maximum likelihood methods, based on data from all type strains of the isolates’ closest relatives. The ITS sequences were aligned using CLUSTAL-X,18 and bootstrap analysis was conducted with 1000 replicates to assess the confidence of the branching.
Phylogenetic analysis
For phylogenetic analysis, the nucleotide sequences were aligned using neighbor-joining algorithm, a multiple sequence alignment program.19 MEGA 7 version software was used to perform phylogenetic based analysis.20 A neighbor-joining algorithm analysis was performed using a heuristic search. An initial tree was generated through stepwise calculation, with sequences added randomly for 1000 replicates. The stability of the tree was assessed with 1000 bootstrap replication.
Qualitative analysis of fungal isolates for production of the ligno-cellulolytic enzymes
Qualitative screening of CMCase and Xylanase
The cellulolytic and xylanolytic capabilities of the fungal strains were assessed using the Congo Red test, as described by Teather and Wood,21 which showed a clearing zone on carboxymethyl cellulose (CMC) agar plates. The CMC and Xylan agar plates were prepared by adding 1% (w/v) CMC and 0.5% (w/v) beechwood Xylan, respectively, to the basal medium, along with 2% agar. The medium was composed of 1.5 g KH2PO4, 5.0 g K2HPO4, 0.5 g (NH4)2SO4, 0.1 g MgSO4.7H2O, 0.2 g NaCl, and 0.1 g yeast extract. The final volume was made up 1 litre by adding distilled water and the pH adjusted to 7.2. The medium underwent sterilisation using autoclaving at a pressure of 15 pounds per square inch for a duration of 20 minutes. The pure selected fungal culture was inoculated into the Xylan and CMC agar containing plates and incubated at 28 °C. Following a five-day incubation period, the plates were immersed in a solution of Congo Red (1 mg/ml in distilled water). After a 20 minute period of incubation, the dye was extracted and the plates were immersed in a solution of 5 M NaCl. After a duration of 15 minutes, the NaCl was removed, and colonies that exhibited the production of CMCase and xylanase were distinguished by the presence of a pale orange to clear region surrounding the colonies in contrast to the red background.
Laccase activity (Modified method of Hankin and Anagnostakis22)
The selected pure fungal culture was inoculated on agar plates containing 2% malt extract and incubated at 28 °C for a period of 7 days. Succeeding this incubation period, a cavity was made in the middle of each plate, and the bottom of the cavity was sealed using molten agar. To evaluate the activity of laccase, a solution of guaiacol (2-methoxyphenol) was prepared by mixing 1 ml of guaiacol with 95% ethanol at a concentration of 1% v/v. Subsequently, 1 ml of this freshly prepared guaiacol solution was added to each cavity, and the plates were kept in darkness. After a period of 10 hours, the plates were examined for the presence of a red to purple colour, which indicates the production of laccase.
Lignin Peroxidase activity (modified method of Egger23)
The selected fungal culture was grown on agar plates containing 2% malt extract and incubated at 28 °C for a period of 7 days. After incubation, molten agar was used to seal the bottom of each plate after a well of 6 to 8 mm in diameter was made there using a sterile cork borer. Each well was filled with 0.5 ml of a 1.0% (w/v) pyrogallol aqueous solution, and the plates were kept in darkness at room temperature for ten hours to assess the activity of lignin peroxidase. The presence of lignin peroxidase activity was indicated by the appearance of a golden yellow to brown hue.
Quantitative analysis of fungal isolates for the production of the lignolytic enzymes
The isolates were grown in 50 ml of sterilised Reese mineral medium with 1% (w/v) rice stubble chopped into 2-3 cm lengths. The flasks were incubated at 30 °C for a duration of 14 days under submerged conditions. After the incubation period, the fermented broth was filtered by using Whatman filter paper No. 41. The filtrate obtained was used to quantitatively measure the activities of Carboxymethyl cellulase (CMCase), Filter Paperase (FPase), xylanase, lignin peroxidase, and laccase.
Determination of Carboxymethyl Cellulase (CMCase) activity
The CMCase quantity in the culture filtrate was measured by using the method described by Ghose.24 The substrate used in the experiment was CMC, obtained from Central Drug House Ltd., Mumbai. Two grams of CMC were added in 100 ml of citrate buffer with a concentration of 0.05 M and a pH of 4.8. After that, a 0.5 ml of enzyme filtrate was taken with 0.5 ml of a solution containing 2% CMC. The mixture was then kept at a temperature of 50 °C for a duration of 30 minutes. The concentration of reducing sugars in each tube was measured using the methodology outlined by Miller.25 An enzyme blank was prepared in a similar manner, excluding the addition of substrate. The enzymatic activity of the filtrate was measured in units per millilitre (U/mL), indicating the amount of enzyme release one microgram of reducing sugars per minute under the submerged conditions.
Determination of Filter Paperase activity (FPase)
The FPase activity or endo β-1,4 glucanase was assessed using the procedure described by Mandels et al.26 The substrate used was Whatman filter paper No. 1, which was cut into pieces of roughly 1 mm. An incubation was performed by combining 50 milligrams of filter paper with 0.5 ml of citrate buffer and 0.5 ml of enzyme filtrate. The solution was incubated at 50 °C for a duration of 60 Minutes. An enzyme solution, devoid of substrate, was made using a similar method. Following the incubation period, the tubes were cooled using flowing tap water. The reducing sugars released in each tube was quantified by the methodology outlined by Miller.25 As per the regulations established by the International Union of Biochemistry, a unit of enzyme activity is defined as the amount of enzyme capable of rescuing one microgram of reducing sugars per minute under submerged conditions.
Quantitative estimation of xylanase activity
A 1% xylan solution was created by dissolving 1 g of xylan in 1 N NaOH and then increasing the volume to 100 ml using 0.05 M citrate buffer at a pH of 4.8. In order to assess the activity of xylanase, test tubes were filled with 0.5 ml of enzyme filtrate and then enriched with a 1% xylan solution. Subsequently, the tubes incubated in water bath set at 50 °C for a period of 30 minutes. The quantification of reducing sugars was conducted using the DNSA method, with absorbance readings taken at a wavelength of 575 nm. A standard curve was generated by employing D-xylose. The definition of one enzyme unit is the liberation of 1 µmole of xylose by hydrolysis. Blanks were created by utilising culture filtrates that had been rendered inactive by the use of heat. The substrate blanks were prepared by combining of citrate buffer (0.5 ml) and of Xylan (0.5 ml) solution at a pH of 4.8.
Quantitative estimation of lignin peroxidase activity
The lignin peroxidase enzyme quantified by using the procedure outlined by Tien and Kirk.27 According to this, add 0.5 ml of enzyme filtrate into cuvettes that already contained 0.05 M of citrate buffer (0.5 ml) at a pH of 4.8. By this combination, 50 µl of a 0.05 M solution of Azure-B and 50 µl of a 10 mM solution of H₂O₂ were introduced. A blank was made without the inclusion of hydrogen peroxide (H₂O₂). The absorbance was recorded at a wavelength of 651 nm at a interval of 30-second for a total period of 180 seconds. A change in absorbance of 0.01 unit at a wavelength of 651 nm per millilitre per minute was deemed to be equal to one unit of enzyme activity.
Quantitative estimation of laccase activity
The laccase activity was determined by measuring the change in absorbance at 436 nm using 5 mM ABTS as the substrate. This was done following the methodology described by Munoz et al.28 In which, add 1 ml enzyme filtrate was mixed with 1 ml of a 0.05 mM citrate buffer (pH 4.8) in a cuvette. In order to start the reaction, a volume of 0.2 ml of a 5 mM solution of ABTS was introduced into the cuvette that already contained the sample. The blank did not undergo the addition of ABTS. Absorbance measurements were taken at a wavelength of 436 nm every 30 seconds for a total of 180 seconds. A laccase unit was determined as a change of 0.01 units in absorbance at 436 nm per millilitre.
Primary screening of filamentous fungi
The current study aims to investigate potent filamentous fungi for selective cellulolytic and ligninolytic activities. During screening 39 (F1 to F39) different morphologically distinct isolates screened for different enzymatic activity. All strains were subjected to preliminary screening.
Identification of lignocellulolytic fungi
Morphological observations and identification
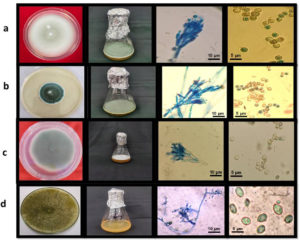
Morphological characteristics such as size, septation, shape, and color of conidiophores and conidia were examined. Based on these morphological features, the isolates were found to exhibit significant differences (Table 1, Figure 1).
Table (1):
Morphological confirmation of lignocellulolytic fungal Strains
Groups |
No. of isolates |
Cultural and morphological characters |
|
|---|---|---|---|
Group 1 |
F2, F3, F4, F26 |
Trichoderma reesei (F-26) (Figure 1d) |
Trichoderma reesei displayed transparent and yellow mycelium growth throughout the plate, as illustrated in Figure 3, which showcases all pertinent characteristics. Several faint white conidia were observed to grow slightly around the periphery. The media displayed a yellow hue. The fully developed culture of T. reesei exhibited the existence of concentric rings either 1 or 2, with one encircling the point of inoculation and second located at the outside edge. This fungus displayed elongated and sinuous branches. The phialides of this fungus frequently displayed a morphology like a flask-shaped, with a swelling base, and were not tightly grouped together. Phialides were commonly organised in pairs. The morphology of conidia exhibited an ellipsoidal shape. |
Group 2 |
F5, F6, F7, F8, F9, F10, F11, F12, F13, F14, F15, F16, |
Talaromyces pinophilus (F-12) (Figure 1b) |
The colonies exhibit moderate growth, reaching a diameter of around 2.5 to 2.8 cm after a span of 7 days. They have a funiculose appearance, characterized by a basal felt with Pure Yellow aerial mycelium. After 10 days, the colonies generate many ascomata on the felt, which are either Light Yellow or Pure Yellow in colour. Conidiogenesis is infrequent and not easily noticeable. Exudates are scarce. Ascomata are typically spherical, with a diameter of 100-300 µm, and can be either Sulphur Yellow or Pure Yellow. They may appear individually or merge together, and reach maturity within a span of 10 to 14 days. The covering is made up of delicate interwoven networks of hyphae, which are surrounded by loose wefts of yellow, densely encrusted, somewhat twisted hyphae that mainly radiate outward. The cleistothecia are small, round to slightly round structures that are found on the surface. They do not have an opening and are generally yellow in color. The asci, which are the reproductive structures, are broadly ellipsoidal to slightly round in shape. The ascospores are ellipsoidal in shape, with some strains being slightly bigger. They have a very fragile surface covered in small spines, which are typically randomly arranged. |
Group 3 |
F17, F18, F19, F20, F21, F22, F23, F24, |
Penicillium griseofulvum(F-22) (Figure 1c) |
Penicillium griseofulvum typically forms colonies on agar media with greenish-grey hues, often exhibiting a velvety or powdery texture. Microscopically, it features conidiophores bearing chains of conidia. Notably, its conidia are smooth-walled and typically round or oval-shaped. The phialides, responsible for conidia production, are flask-shaped. This species is characterized by its ability to synthesize griseofulvin, a bioactive compound with antimicrobial properties. Its cultural and morphological traits, alongside its metabolic capabilities, make Penicillium griseofulvum of significant interest in pharmaceutical and biotechnological research, particularly in the study of natural product synthesis and microbial ecology. |
Group 4 |
F25, F1, F27, F28, F29, F30, F31, F32, F33, F34, F35 F36, F37, F38, F39, |
Penicillium oxalicum (F-1) (Figure 1a) |
The colonies on CYA have a diameter of 20-30. They are plain or lightly radially grooved, and have a fluffy to very deeply fluffy texture. The margins are deep, with white mycelium at the edges. In the centre, they are yellow to pale orange or buff in colour. Conidiogenesis is light, with a grey green or inconspicuous appearance. Exudates are usually present and can be clear to brown in color. The colonies typically produce a brown to reddish brown soluble pigment. The reverse side of the colonies is brown or slightly greyer than amber, although some isolates may have less intense pigmentation. Conidiophores emerge from either surface or aerial hyphae, with lengthy stipes. The walls of the structure are either smooth or have a very fine rough texture. The structure has terminal groups of 2-4 small branches, and each branch has 5-8 closely packed, flask-shaped structures with short extensions. The spores produced by this structure are spherical in shape. |
Figure 1. Morphological observations and identification of a: Penicillium oxalicum (F-1); b: Talaromyces pinophilus (F-12); c: Penicillium griseofulvum (F-22); d: Trichoderma reesei (F-26)
Molecular identification of the promising lignocellulolytic fungal isolates
The promising strains based on qualitative and quantitative lingo-cellulolytic screening underwent molecular confirmation. The morphological identification was confirmed through, molecular characterizations using Internal transcribed spacer (ITS) sequences. PCR based amplification of all the isolates with both forward and reverse primers of ITS (ITS1 and ITS4) were carried out. Amplified products were separated and sequenced. Further the amplified products were sent to sequencing. After receiving the sequencing result, sequences were analysed for their quality through Bio-edit software. The sequences from the amplified product were blasted with NCBI GenBank database (webpage: http://blast.ncbi.nlm.nih.gov) comparing with the reference sequences available at NCBI GenBank database. Similarity between 97 to 100 percent was obtained (Table 2).
Table (2):
Molecular identification of screened fungal strains confirmation
Name |
Maximum similarity |
Sequence similarity with NCBI database through BLAST analysis |
Accession no. |
|---|---|---|---|
Penicillium oxalicum (F1) |
Penicillium oxalicum |
97% |
PQ276969 |
Talaromyces pinophilus (F12) |
Talaromyces pinophilus |
98% |
PQ276970 |
Penicillium griseofulvum (F22) |
Penicillium griseofulvum |
99% |
PQ276971 |
Trichoderma reesei (F26) |
Trichoderma reesei |
100% |
PQ276972 |
Phylogenetic analysis
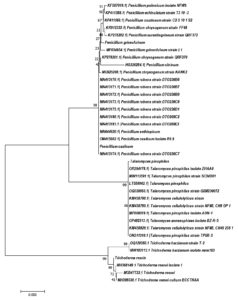
A phylogenetic study was conducted utilising the neighbor-joining technique. The results of the analysis revealed that all four isolates (F1, F12, F22, F26) demonstrated the highest similarity with their Ex-type strains retrieved from NCBI GenBank, compared to the other Penicillium and Trichoderma species (Figure 2), including Penicillium polonicum, Penicillium echinulatum, Penicillium crustosum, Penicillium chrysogenum, Penicillium aurantiogriseum, Penicillium citrinum, Penicillium ruben, Penicillium aethiopicum, Talaromyces cellululolyticum, Talaromyces annesophieae and Trichoderma harzianum.
Figure 2. Phylogenetic tree of four strains created using neighbor-joining algorithm with 1000 replications
Qualitative screening of CMCase and Xylanase
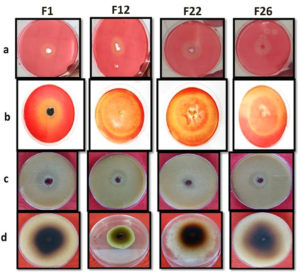
Thirty-nine fungal isolates were isolated from the ICAR-IARI paddy fields in New Delhi, India, and screened for maximum activities, cellulolytic, xylanolytic, laccase, and lignin peroxidase enzymatic activities. The initial screening of fungal species conducted via observing the development of a distinct and visible zone around the colony on the solid media, which was reinforced by particular indicators. The formation of a pale-yellowish hydrolyzed zone on solid media when Congo red dye is used indication of extracellular CMCase as well as xylanase production by filamentous fungi. In this study found that, the formation of smaller fragment that do not effectively bind Congo red dye, thereby causing the formation of a pale-yellowish hydrolyzed zone around the fungal colonies. This qualitative assay serves as an indication of the extracellular secretion of CMCase and xylanase by filamentous fungi (Figure 3a, 3b).
Qualitative screening of Laccase activity
Qualitative
When guaiacol is employed as the substrate, the presence of a transparent patch over solid media shows that filamentous fungus is producing laccase outside of its cells. Therefore, the lack of colour in the area surrounding the fungal colony signifies the existence of laccase activity, as it facilitates the chemical reaction that converts guaiacol into a substance without colour. When guaiacol is utilised as a substrate, a distinct zone appears on the solid media, indicating that filamentous fungus are producing extracellular laccase (Figure 3c).
Qualitative screening of Lignin Peroxidase activity
Selected strain plates will be filled with 0.5 ml of a 1.0% (w/v) pyrogallol aqueous solution, and the plates were kept in darkness at room temperature for ten hours to assess the activity of lignin peroxidase. The presence of lignin peroxidase activity was indicated by the appearance of a golden yellow to brown hue (Figure 3d).
Figure 3. Primary screening results of fungal strain for: a: CMCase activity; b: Xylanase activity; c: Laccase activity; d: Lignin peroxidase activity
Quantitative estimation
Assay of Cellulase Enzyme Production
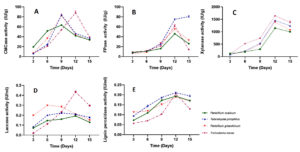
The four selected fungal strains Penicillium oxalicum, Talaromyces pinophilus, Penicillium griseofulvum, and Trichoderma reesei were screened for qualitative hydrolytic potential. The hydrolytic potential of fungi was estimated with time under submerged fermentation using chopped paddy stubble as the substrate. Table 3 shows CMCase, FPase and xylanolytic activity of all fungal strains 3, 6, 9, 12 and 15 days of submerged Fermentation. The data obtained for CMCase activity shows (Figure 4a) that in case of Penicillium oxalicum, Talaromyces pinophilus and Penicillium griseofulvum the enzyme activity increased from 3 days to 12 days showing 19.475 IU/g, 6.25 IU/g and 6.93 IU/g after three days of incubation and increased to maximum on 9th day with 63.424 IU/g, 83.72 IU/g and 82.78 IU/g respectively. Among all the four strains Trichoderma reesei resulted as maximum hydrolytic potential 88.26 IU/g on 15th day after inoculation and incubation.
Table (3):
Quantitative estimation of cellulolytic and hemicellulolytic potential of selected four fungal strains under submerged Fermentation
| Strain | CMCase (IU/g) | FPase (IU/g) | Xylanase (IU/g) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (Days) | Time (Days) | Time (Days) | |||||||||||||
| 3 | 6 | 9 | 12 | 15 | 3 | 6 | 9 | 12 | 15 | 3 | 6 | 9 | 12 | 15 | |
| F1 | 19.475 ± 0.012d | 51.43 ± 0.057a | 63.424 ± 0.032a | 42.012 ± 0.021b | 33.77 ± 0.012c | 8.342 ± 0.020d | 10.7 ± 0.015c | 16.11 ± 0.025b | 46.01 ± 0.035d | 26.28 ± 0.026a | 121.7 ± 0.026d | 195.24 ± 0.032c | 301.44 ± 0.049b | 1146.1 ± 0.038a | 998.78 ± 0.079a |
| F12 | 6.25 ± 0.075d | 24.59 ± 0.026c | 83.72 ± 0.030d | 46.65 ± 0.026a | 36.79 ± 0.035b | 7.803 ± 0.023d | 11.032 ± 0.023c | 25.33 ± 0.026b | 74.94 ± 0.026a | 80.66 ± 0.019a | 82.14 ± 0.039d | 229.57 ± 0.012c | 532.55 ± 0.052b | 1458.2 ± 0.018a | 1238.25 ± 0.020a |
| F22 | 6.93 ± 0.012d | 36.93 ± 0.021b | 82.78 ± 0.021a | 43.84 ± 0.038a | 32.97 ± 0.015c | 6.27 ± 0.021d | 8.47 ± 0.021c | 26.99 ± 0.024b | 55.1 ± 0.026a | 32.89 ± 0.036a | 92.78 ± 0.026d | 190.77 ± 0.035c | 498.63 ± 0.035b | 1383.12 ± 0.018d | 1065.9 ± 0.046a |
| F26 | 6.52 ± 0.032d | 20.99 ± 0.061c | 38.93 ± 0.030b | 53.87 ± 0.038a | 88.26 ± 0.030c | 9.14 ± 0.023d | 11.73 ± 0.038c | 21.41 ± 0.035a | 61.82 ± 0.044b | 14.06 ± 0.044b | 112.41 ± 0.062d | 521.63 ± 0.058c | 744.28 ± 0.026b | 1640.52 ± 0.015d | 1378.98 ± 0.086a |
Assay of FPase enzyme production
Further FPase activity data reveals (Table 3, Figure 4b) that Penicillium oxalicum, Penicillium griseofulvum and Trichoderma reesei fungal strain shows maximum potential on 12th day having 46.01 IU/g, 55.1 IU/g and 61.82 IU/g respectively, while Talaromyces pinophilus showed maximum hydrolytic potential on 15th day having 80.66 IU/g and this fungal strain showed the highest potential among the four selected strains.
Assay of xylanase enzyme production
Similarly, xylanase activity data reveals (Figure 4c) that all four fungal strains Penicillium oxalicum, Talaromyces pinophilus, Penicillium griseofulvum and Trichoderma reesei showed maximum enzymatic activity on 12th day showing 1146.1 IU/g, 1458.2 IU/g, 1383.12 IU/g and 1640.52 IU/g, respectively (Table 3). Among all the four fungal strains Trichoderma reesei showed the highest xylanase activity followed by Talaromyces pinophilus, Penicillium griseofulvum and Penicillium oxalicum fungal strains.
Assay of laccase enzyme production
Since lignolytic and hydrolytic potential has to be estimated, all four fungal strains were estimated for Laccase and lignin peroxidase activity under submerged Fermentation condition. Table 4 shows the hydrolytic activities of four strains with time in days. The fungal strains showed initial increase in Laccase activity (Figure 4d) from 3rd day to 9th and 12th day. The maximum laccase activity was observed by strain Trichoderma reesei on 12th day showing 0.434 IU/ml which was followed by strain Penicillium griseofulvum on 9th day showing 0.287 IU/ml; Talaromyces pinophilus also showed maximum activity on 9th day showing 0.223 IU/ml while the strain Penicillium oxalicum showed 0.193 IU/ml on 12th day. The data clearly shows variation in hydrolytic potential of all four fungi with time. However, overall observation indicates Trichoderma reesei fungal strain has potential for lignolytic activity.
Table (4):
Quantitative estimation lignolytic potential of selected four fungal strains under submerged Fermentation
| Strain | Laccase activity (IU/ml) | Lignin peroxidase activity (IU/ml) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time (days) | Time (days) | |||||||||
| 3 | 6 | 9 | 12 | 15 | 3 | 6 | 9 | 12 | 15 | |
| F1 | 0.072 ± 0.007c | 0.147 ± 0.012b | 0.161 ± 0.008ab | 0.193 ± 0.008a | 0.129 ± 0.006b | 0.055 ± 0.008c | 0.073 ± 0.009c | 0.109 ± 0.005b | 0.171 ± 0.007a | 0.172 ± 0.006a |
| F12 | 0.083 ± 0.005a | 0.2 ± 0.033a | 0.223 ± 0.006a | 0.21 ± 0.070a | 0.178 ± 0.008a | 0.042 ± 0.007c | 0.094 ± 0.010b | 0.143 ± 0.009b | 0.194 ± 0.007a | 0.187 ± 0.009a |
| F22 | 0.199 ± 0.008ab | 0.3 ± 0.065a | 0.287 ± 0.006a | 0.212 ± 0.005ab | 0.152 ± 0.011b | 0.045 ± 0.006c | 0.113 ± 0.006b | 0.119 ± 0.007b | 0.169 ± 0.005a | 0.151 ± 0.009a |
| F26 | 0.02 ± 0.005c | 0.112 ± 0.007b | 0.23 ± 0.047a | 0.434 ± 0.007c | 0.297 ± 0.007a | 0.066 ± 0.008a | 0.057 ± 0.005a | 0.07 ± 0.044a | 0.103 ± 0.005a | 0.129 ± 0.004a |
Assay of lignin peroxidase enzyme production
Similar trends were found in case of lignin peroxidase activity (Figure 4e) and there was marginal difference among the four strains. The two fungal strains Penicillium oxalicum and Trichoderma reesei showed maximum lignin peroxidase activity 0.172 IU/ml and 0.129 IU/ml on 15th day while strain Talaromyces pinophilus and Penicillium griseofulvum showed maximum activity on 12th day having a 0.194 and 0.169 IU/ml, respectively (Table 4). These results give an insight for lignolytic potential of all the four fungi where it is found that that all are potential candidates for this desirable activity. On the whole, all four fungi are showing hydrolytic potential and can be considered for microbial consortium development formulation.
A total of fifteen fungi were isolated for their xylanase and cellulase activity with an aqueous solution (1%) of Congo red dye. After, 7-days of incubation period at 50 °C, the culture plates were fully flooded with Congo red dye solution and allowed to settle undisturbed for a period of 30 minutes. Then, the destaining process was performed by washing the culture plates twice with a 1 M NaCl solution for 20 minutes each. The study by Ahirwar et al.29 identified transparent hydrolytic zones, indicating the decomposition of cellulose and xylan into basic sugars.
There were 23 isolates exhibiting distinct zones on xylan containing agar plates. The fungal morphology and xylanase activity was done on potato dextrose agar and agar plates containing the xylan as a substrate, respectively and found the 5 fungal isolates that exhibited the highest enzymatic activity, as reported by Dhaver et al.30 Among the 22 fungal strains tested for xylanase activity, a positive response was observed, with clear zones indicating producer strains. However, quantitative analysis is necessary to identify the highest producers of xylanase.31 Plates (a) and (b) display zones of xylan hydrolysis, while plates (c) and (d) show zones of mannan hydrolysis.
Ahirwar et al.29 reported similar findings, observing that Absidia corymbifera CM-4, A. fumigatus SS-1, Humicola insolens WS-20, T. lanuginosus SL-9, M. thermophila NFCCI 3725, E. nidulans WS-23, Aspergillus terreus SS-43, C. thermophile L-61, Scytalidium thermophile SL-36, M. albomyces DOM-65 and M. cinnamomea NFCCI 3724 exhibited clear zones against an opaque xylan agar medium.
The most frequently employed microbes for hydrolytic enzyme production are typically from the genera Penicillium, Aspergillus, and Trichoderma.32 Phanerochaete chrysosporium, a white rot fungus, has been shown in study to produce significant quantities of cellulase enzyme.33 Recent research indicates that specific types of white rot fungus have great potential for breaking down biomass and removing lignin due to their ability to create enzymes that can break down cellulose, hemicellulose, and lignin.34 The size of the hydrolysis zone was found to be positively correlated with the diameter of the colony on CMC and xylan agar mediums. However, all of the isolates exhibited slightly higher enzymatic activity.35,36
Seventeen fungal isolates exhibited cellulolytic activity by their interaction with Congo red, which specifically binds to unbroken β-D-glucan present in Carboxymethyl cellulose. The areas where cellulose was broken down via the enzymes were visible as transparent zones or faint rings.37 In a prior investigation by Su et al.,38 63 fungal strains exhibiting lignolytic properties were isolated, indicating that fungal isolates were more proficient in lignin peroxidase activity compared to laccase activity.39 discovered those ten out of twenty-one isolated cultures tested positive for laccase, manifesting a reddish-brown zone on petri plates.40 Screened ten fungal strains for ligninolytic enzyme production using ABTS and Azure B dye decolorization. Fungal strains such as A. ochraceus, Rhizopus sp., F. africana, F. verticillioides, A. nomius, and A. favus exhibited clearance zones indicating lignin degradation, as did P. pulmonarius and Fomitopsis sp. A subsequent evaluation was carried out to analyse the formation of extracellular laccase employing guaiacol as a phenolic substrate.41 Nayak and Choudhary42 utilised a comparable methodology using the guaiacol plate test for the purpose of qualitative screening. Prior research has revealed many fungi with lignolytic characteristics in natural habitats.38,43 Thrimothi et al. and Nayak et al.39,44 conducted a qualitative assessment of 23 lignolytic fungal isolates with the guaiacol test on PDA plates. They identified 5 fungal isolates that had very strong laccase activity.
Similarly, Devi et al.31 quantified cellulolytic fungal strains under submerged fermentation using abundant and inexpensive rice stubble. Notable activity was observed 0.92 U/m for Aspergillus flavus and 0.70 U/mL for Aspergillus terreus. The study conducted by Septiani et al.45 investigated cellulase production utilising CMC media. The results revealed that the crude extract of Aspergillus niger demonstrated a (0.131 U/mL) cellulase activity, whereas Trichoderma reesei exhibited a cellulase activity of 0.106 U/mL. Andriani et al.46 performed a quantitative analysis of cellulase activity using the DNS method, quantifying Trametes hirsuta at 540 U/L after 12 days of incubation. Intasit et al. (2021) reported cellulase activity for Aspergillus tubingensis TSIP9 (40.09 U/g), Aspergillus niger ATCC 6815 (37.31 U/g), and Trichoderma reesei QM 9414 (9.26 U/g).
Giwa et al.47discovered that Trichoderma orientalis is a notable cellulase producer, along with other filamentous fungi like Aspergillus sp., Trichoderma sp., and white rot fungi, specifically Phanerochaete chrysosporium.48 Sakpetch et al.49 evaluated the production of cellulase enzyme by three isolates (F6, A2, and B15). They discovered that the CMCase activities were F6 (0.61 U/ml), A2 (0.33 U/ml), and B15 (0.21 U/ml).50 Conducted a study which found that isolate HST16 had the highest CMCase activity at a concentration of 0.026 IU/ml. The CMCase activities of Aspergillus terreus and Aspergillus flavus were found to be the lowest 0.179 U/ml and highest 2.03 U/ml, respectively, wheat straw was used as a carbon source. The CMCase activity on rice straw were measured by A. terreus of 1.07 U/ml.31 Lee et al.51 examined the Filter paperase activity of 64 fungal isolates, measuring activities that varied from 0 to 0.259 U/ml. The FPase activities of specific isolates were measured as follows: Aspergillus flavus displayed an activity of 1.169 U/ml, Ramichloridium apiculatum exhibited 1.03 U/ml activity, and another Aspergillus flavus isolate exhibited 0.99 U/ml activity. The Filter paperase activity in rice stubble varied between 0.163 and 1.07 U/ml. The RT isolate exhibited an FPase activity (0.512 U/ml), while the AL1 white isolate displayed an FPase (0.174 U/ml) activity.31,50 Devi et al. also documented an FPase activity of 0.026 IU/ml for the HST16 isolate.31 Devi et al. evaluated the xylanolytic activity of fungal strains, both isolated and commercial, utilising wheat straw and rice stubble in a submerged fermentation process. Significant xylanase activity was detected in Aspergillus flavus (0.92 U/ml) and Aspergillus terreus 0.70 U/ml. According to Andriani et al,46 Trametes hirsuta exhibited a xylanase activity of 670 U/L after being incubated for 10 days.
All the strains (Aspergillus terreus, Aspergillus flavus, A. terreus ITCC 11853.23, Aspergillus flavus ITCC 11854.23, RT isolate) producing xylanase on wheat straw exhibited activities ranging from 0.33 to 4.03 U/ml. Among these, Aspergillus terreus demonstrated the highest xylanolytic activity at 4.03 U/ml under submerged fermentation. Additionally, significant xylanase production was observed in Aspergillus flavus (2.81 U/ml), A. flavus ITCC 11854.23 found 2.96 U/ml, and Aspergillus terreus ITCC 11853.23 found 2.28 U/ml. The xylanase activity exhibited by the isolated fungal strains on rice straw showed variation within a range of 0.321 to 4.68 U/ml, with Aspergillus flavus ITCC 11854.23 showing the highest activity at 4.68 U/ml. Additionally, the study by Devi et al.31 found Aspergillus terreus ITCC 11853.23 (2.64 U/ml), the RT isolate (1.70 U/ml), and Aspergillus flavus ITCC 11694.22 (1.83 U/ml) as highly proficient in producing xylanase.
The xylanase activity exhibited by the isolated fungal strains on rice stubble showed variation within a certain range. When evaluating xylanase activity in three fungal strains, A. niger ATCC 6815 exhibited markedly greater xylanase activity (82.89 U/g) compared to T. reesei QM 9414 (40.97 U/g) and A. tubingensis TSIP9 (62.43 U/g).3 Dhaver et al.30 conducted a study where they measured the xylanase activities in different strains. They found that strain CB1 had an activity of 21.67 U/ml, CB2 had 16.98 U/ml, MS5 had 22.98 U/ml, PS3 had 15.64 U/ml, and PB7 had 14.22 U/ml. Tanwar et al.52 discovered that among the 10 fungal isolates tested for xylanase activity, 5 fungal isolates (SH2, SH5, R3, R4, and S5) demonstrated the most elevated activities, measuring 7.20, 7.44, 7.31, 7.83, and 8.09 IU/ml, respectively. Furthermore, Olanbiwoninu and Odunfa53 documented that the fungus Aspergillus terreus produced xylanase enzyme of 3.4 U/ml. In their study, Bisht et al.54 specifically chose 12 fungal isolates to produce enzymes in liquid broth medium containing degraded bark as the substrate. They revealed that three isolates (A2, B15 and F6,) exhibited the highest enzymatic activity, measuring at 0.36, 0.32 and 0.61, IU/ml, respectively.
The observations we made are supported by the findings of Ang et al.43 They found that white-rot fungus, notably Penicillium sanguineus, Penicillium chrysosporium, and Penicillium radiata, possess ligninolytic activities and are capable of breaking down lignin and compounds similar to lignin. Furthermore, our analysis confirmed the presence of laccase and lignin peroxidase activities in P. sajor-caju. Sumiati et al.55 made similar findings, measuring the laccase enzyme generated by isolates KRB12 at 8244.72 U/mL, KRB1 at 3239.68 U/mL, and KRB8 at 3243.54 U/mL, Sijinamanoj et al.56 reported that the fungal isolate T. harzianum produced a laccase enzyme at a rate of 0.059 U/mL over a 15-day period. Illuri et al.2 previously reported similar findings, documenting laccase activities for various species as follows: Pleurotus djamor (14.05 × 10-6 IU/mL), Hypsizygus ulmarius (14.83 × 10-6 IU/mL), Tricholomopsis giganteus (10.73 × 10-6 IU/mL), and Volvariella volvacea (10.45 × 10-6 IU/mL), Pleurotus florida (12.11 × 10-6 IU/mL), and Oudemansiella radicata (11.83 × 10-6 IU/mL).
The importance of screening basidiomycetes fungi for ligninolytic organisms is emphasised by increasing evidence that supports their industrial applications and their ability to degrade lignin through ligninolytic enzymes.57-59 They were examined, P. ostreatus, P. djamor, T. giganteus, and H. ulmarius displayed lignin peroxidase activity that varied from 10.06 to 10.64 × 106 IU/ml. The species P. sajorcaju, P. florida, and V. volvacea exhibited laccase activity of 19.56 × 106 IU/ml, 7.31 × 106 IU/ml, and 7.05 × 106 IU/ml, respectively, as reported by Illuri et al.2 According to Thrimothi et al,39 the fungal isolate Pv5 showed LiP activity of 412 U/ml on the 7th day, which then increased to 544 U/ml by the 14th day. Andriani et al.46 recorded the highest levels of Laccase and LiP activity as 25.7 × 103 and 91 U/L, respectively, following an 8-day incubation period. Sanchez-Corzo et al.60 found that Trichoderma longibrachiatum and Trametes sanguinea produced lignin peroxidase enzyme in liquid media containing coffee husk, resulting in yields of 540.12 U/L and 7115.22 U/L, respectively. On the 12th day, Bisht et al.54 reported peak LiP production at 10.386 U/mL. Olanbiwoninu and Odunfa53 made similar observations, where isolates KRB8 produced 6626.31 U/mL of lignin peroxidase, KRB9 produced 5302.89 U/mL, and KRB1 produced 3881.91 U/mL.
In summary, this study isolated 39 fungal strains from the ICAR-IARI paddy fields in New Delhi, India, and screened them for cellulolytic, xylanolytic, laccase, and lignin peroxidase activities. Four isolates (F1, F12, F22, and F26) demonstrated the highest enzymatic activities and were selected for further investigation. The research evaluated the potential of lignocellulosic residues, specifically rice stubble, as substrates for cultivating these fungi under submerged fermentation at 30 °C over a 15-day period. With rice stubble accounting for 40-60% of the rice plant’s biomass and containing cellulose (42.14%), hemicellulose (22.08%), and lignin (11.98%), it was shown to be a promising renewable resource for energy and biochemical production. The selected fungal strains, Penicillium oxalicum (F1), Talaromyces pinophilus (F12), Penicillium griseofulvum (F22), and Trichoderma reesei (F26) were evaluated for their lignocellulolytic enzyme production. Peak enzyme activities were recorded at 3-day intervals, with CMCase (63.42-88.26 U/mL), FPase (46.01-80.66 U/mL), xylanase (1146.10-1640.52 U/mL), lignin peroxidase (0.192-0.287 U/mL), and laccase (0.193-0.434 U/mL) demonstrating significant potential. ITS sequencing confirmed the identities of the fungal strains. Overall, these findings highlight the potential of these fungi to produce hydrolytic enzymes and efficiently degrade lignocellulose, with rice stubble serving as a cost-effective carbon source for biotechnological applications in biorefineries.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Raymond P, Mshandete AM, Kivaisi AK. Enzyme profiles of Pleurotus HK-37 during mycelia vegetative growth and fruiting on solid sisal waste fractions supplemented with cow manure. Adv Biochem. 2015;3(5):57-65.
Crossref - Illuri R, Kumar M, Eyini M, et al. Production, partial purification and characterization of ligninolytic enzymes from selected basidiomycetes mushroom fungi. Saudi J Biol Sci. 2021;28(12):7207-7218.
Crossref - Intasit R, Cheirsilp B, Suyotha W, Boonsawang P. Synergistic production of highly active enzymatic cocktails from lignocellulosic palm wastes by sequential solid state-submerged fermentation and co-cultivation of different filamentous fungi. Biochem Eng J. 2021;173:108086.
Crossref - Michelin M, Ximenes E, de Moraes Polizeli MDLT, Ladisch MR. Effect of phenolic compounds from pretreated sugarcane bagasse on cellulolytic and hemicellulolytic activities. Bioresour Technol. 2016;199:275-278.
Crossref - Asgher M, Shaha WK, Bilal M. Optimization of lignocellulolytic enzyme production by Pleurotus eryngii WC 888 utilizing agro-industrial residues and bio-ethanol production. Rom Biotechnol Lett. 2016;21:11133-11143.
- Toushik SH, Lee KT, Lee JS, Kim KS. Functional applications of lignocellulolytic enzymes in the fruit and vegetable processing industries. J Food Sci. 2017;82(3):585-593.
Crossref - Hiloidhari M, Das D, Baruah DC. Bioenergy potential from crop residue biomass in India. Renew Sustain Energy Rev. 2014;32:504-512.
Crossref - Bhuvaneshwari S, Hettiarachchi H, Meegoda JN. Crop residue burning in India: Policy challenges and potential solutions. Int J Environ Res Public Health. 2019;16(5):832.
Crossref - Malik K, Sushil NK. Fermentation of paddy straw and fruit wastes for bioethanol production. Int J Chem Stud. 2019;7(3):1756-1759.
- Harun SN, Hanafiah MM, Noor NM. Rice Straw Utilisation for Bioenergy Production: A Brief Overview. Energies. 2022;15:5542.
Crossref - Amer A, Bibi A. Fungal cellulase; production and applications: minireview. LIFE: International Journal of Health and Life Sciences. 2018;4(1):19-36.
Crossref - Reese ET, Mandels M Enzymatic hydrolysis of cellulose and its derivatives. Methods in Carbohydrate Chemistry. 1963;3(2):139-142.
Crossref - Rifai MA. A revision of the genus Trichoderma. Mycological Papers. 1969;116:1-54.
- Bissett J. A revision of the genus Trichoderma. II. Infrageneric classification. Canadian Journal of Botany. 1991;69(11):2357-2372.
Crossref - Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531-6535.
Crossref - White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A guide to methods and applications/Academic Press, Inc. 1990;315-322.
Crossref - Kim OS, Cho YJ, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt_3):716-721.
Crossref - Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876-4882.
Crossref - Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-1797.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43(4):777-780.
Crossref - Hankin L, Anagnostakis SL. The use of solid media for detection of enzyme production by fungi. Mycologia.1975;67(3):597-607.
Crossref - Egger KN. Substrate hydrolysis patterns of post-fire ascomycetes (Pezizales). Mycologia. 1986;78(5):771-780.
Crossref - Ghose TK. Measurement of cellulase activities. Pure & Appl Chem. 1987;59(2):257-268.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426-428.
Crossref - Mandels M, Andreotti R, Roche C. January. Measurement of saccharifying cellulase. In Biotechnol. Bioeng. Symp.; (United States) (Vol. 6). Army Natick Development Center, MA. Biotechnology and Bioengineering Symposium. 1976;6:21-23.
- Tien M, Kirk TK. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238-249).
Crossref - Munoz C, Guillen F, Martinez AT, Martinez MJ. Induction and characterization of laccase in the ligninolytic fungus Pleurotus eryngii. Curr Microbiol. 1997;34:1-5.
Crossref - Ahirwar S, Soni H, Prajapati BP, Kango N. Isolation and screening of thermophilic and thermotolerant fungi for production of hemicellulases from heated environments. Mycology. 2017;8(3):125-134.
Crossref - Dhaver P, Pletschke B, Sithole B, Govinden R. Isolation, screening, preliminary optimisation and characterisation of thermostable xylanase production under submerged fermentation by fungi in Durban, South Africa. Mycology. 2022;13(4):271-292.
Crossref - Devi A, Singh A, Kothari R. Fungi based valorization of wheat straw and rice straw for cellulase and xylanase production. Sustain Chem Environ. 2024;5:100077.
Crossref - Manisha, Yadav SK. Technological advances and applications of hydrolytic enzymes for valorization of lignocellulosic biomass. Bioresour Technol. 2017;245:1727-1739.
Crossref - Manavalan T, Manavalan A, Heese K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr Microbiol. 2015;70:485-498.
Crossref - Xu X, Lin M, Zang Q, Shi S. Solid state bioconversion of lignocellulosic residues by Inonotus obliquus for production of cellulolytic enzymes and saccharification. Bioresour Technol. 2018;247:88-95.
Crossref - Lalita, Prasher IB. Qualitative screening of lignocellulolytic enzymes in wood rotting agaricomycetes from North Western Himalayas. Journal of Advanced Botany and Zoology. 2014;1(3):1-3.
- Bairagi S. Isolation, screening and selection of fungal strains for potential cellulase and xylanase production. Int J Pharm Sci Invent. 2016;5(3):1-6.
- Namnuch N, Thammasittirong A, Thammasittirong SNR. Lignocellulose hydrolytic enzymes production by Aspergillus flavus KUB2 using submerged fermentation of sugarcane bagasse waste. Mycology. 2021;12(2):119-127.
Crossref - Su Y, Yu X, Sun Y, Wang G, Chen H, Chen G. Evaluation of screened lignin-degrading fungi for the biological pretreatment of corn stover. Sci Rep. 2018;8(1):p.5385.
Crossref - Thrimothi D, Sujatha E, Swetha KG, Krishna G. Isolation, Screening, Identification, and Assessment of Laccase-Producing Fungi Isolated From Different Environmental Samples. Biosci Biotechnol Res Asia. 2023;20(4):1303-1315.
Crossref - Civzele A, Stipniece-Jekimova AA, Mezule L. Fungal ligninolytic enzymes and their application in biomass lignin pretreatment. J Fungi. 2023;9(7):780.
Crossref - Saroj P, Manasa P, Narasimhulu K. Characterization of thermophilic fungi producing extracellular lignocellulolytic enzymes for lignocellulosic hydrolysis under solid-state fermentation. Bioresourc Bioprocess. 2018;5(1):1-14.
Crossref - Nayak B, Choudhary R. Optimization, purification and characterization of laccase from lignocellulolytic fungi Dichotomopilus funicola NFCCI 4534 and Alternaria padwickii NFCCI 4535. Biocatal Agric Biotechnol. 2022;42:102344.
Crossref - Ang TN, Ngoh GC, Chua ASM. A quantitative method for fungal ligninolytic enzyme screening studies. Asia-Pacific J Chem Eng. 2011;6(4):589-595.
Crossref - Nayak B, Choudhary R, Roymom MG. Lignocellulolytic Fungal Isolation and Screening for Their Laccase Producing Ability. Indian J Sci Res. 2017;13(2):188-191.
- Septiani DIA, Suryadi H, Munim A, Mangunwardoyo W. Production of cellulase from Aspergillus niger and Trichoderma reesei mixed culture in carboxymethylcellulose medium as sole carbon. Biodiversitas Journal of Biological Diversity. 2019;20(12).
Crossref - Andriani A, Maharani A, Yanto DHY, et al. Sequential production of ligninolytic, xylanolytic, and cellulolytic enzymes by Trametes hirsuta AA-017 under different biomass of Indonesian sorghum accessions-induced cultures. Bioresour Technol Rep. 2020;12:100562.
Crossref - Giwa AS, Ali N, Akhter MS. Cellulose Degradation Enzymes in Filamentous Fungi, A Bioprocessing Approach Towards Biorefinery. Mol Biotechnol. 2023:1-15.
Crossref - Faison BD, Kirk TK. Relationship between lignin degradation and production of reduced oxygen species by Phanerochaete chrysosporium. Appl Environ Microbiol. 1983;46(5):1140-1145.
Crossref - Sakpetch P, Aran H, Chandumpai A. Isolation and Screening of potential lignocellulolytic microorganisms from rubber bark and other agricultural residues. Walailak J Sci Technol. 2017;14(12):953-967.
- Tanvi SC, Dhanker R, Goyal S. Assessment of the potential of promising fungal isolates for Lignocellulosic biomass utilization under controlled conditions. Pharma Innovation Journal. 2018;7(8):532-537
- Lee H, Lee YM, Heo YM, Lee J, Kim JJ. Evaluation of Cellulolytic Enzyme Production by Indigenous Fungi in Korea. Korean J Environ Biol. 2017;35(4):648-653.
Crossref - Tanwar D, Sharma N, Sharma N, Prusty PK. Isolation and screening of fungi from rotten wood for various hydrolytic enzymes production. Ann Phytomed. 2020;9(1):122-128.
Crossref - Olanbiwoninu AA, Odunfa SA. Production of cellulase and xylanase by Aspergillus terreus KJ829487 using cassava peels as subtrates. Adv Microbiol. 2016;6(7):502-511.
Crossref - Bisht M, Rathi N, Rai JPN. Importance of lignin modifying enzymes from isolated white rot fungus in lignin degradation. Int J Innov Res Sci Eng Technol. 2017;6(7):2347-6710.
- Sumiati T, Suryadi H, Harmita, Sutriyo. Isolation of white rot fungi from rotten wood from bogor botanical garden in Indonesia and its ligninolytic enzymes activity. Pharmacognosy Journal. 2022;14(1).
Crossref - Sijinamanoj V, Muthukumar T, Muthuraja R, et al. Ligninolytic valorization of agricultural residues by Aspergillus nomius and Trichoderma harzianum isolated from gut and comb of Odontotermes obesus (Termitidae). Chemosphere. 2021;284:131384.
Crossref - Ire FP, Ahuekwe EF. Production of fungal laccase using orange peelings as substrate by submerged static fermentation. Br Microbiol Res J. 2016;15(5):1-19.
Crossref - Chen H, Liu J, Chang X, et al. A review on the pretreatment of lignocellulose for high-value chemicals. Fuel Process Technol. 2017;160:196-206.
Crossref - Pandey RK, Tewari S, Tewari L. Lignolytic mushroom Lenzites elegans WDP2: Laccase production, characterization, and bioremediation of synthetic dyes. Ecotoxicol Environ Safety. 2018;158:50-58.
Crossref - Sanchez-Corzo LD, Alvarez-Gutierrez PE, Meza-Gordillo R, Villalobos-Maldonado JJ, Enciso-Pinto S, Enciso-Saenz S. Lignocellulolytic enzyme production from wood rot fungi collected in chiapas, mexico, and their growth on lignocellulosic material. J Fungi. 2021;7(6):450.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.