ISSN: 0973-7510
E-ISSN: 2581-690X
The pathogens attained through nosocomial infection exhibit a higher degree of antibiotic resistance due to constant exposure to drug therapy. There is a need to develop alternate therapeutics for treating these resistant pathogens. Objective: The objective of this study is to isolate pathogens from hospital-acquired infection to determine its epidemiology and multidrug resistance. In this study, urine and swab samples (354) were obtained from hospitalized patients with no prior infection history. For screening antibiotic resistance among the isolates, 15 antibiotics were used in this study, and also various piperidine compounds were used to evaluate the minimum inhibitory concentration against the isolates. Among them, 160 reported positive for the presence of Staphylococcus species (37), Salmonella species (23), Pseudomonas species (27), Proteus species (21), E. coli (34) and Klebsiella species (18). Mostly, all the pathogens obtained from clinical cases show high antibiotic resistance. The highest percentage of resistance was recorded against amoxicillin and penicillin (98%). The least rate of resistance was noticed against gentamycin (42%). Like antibiotics, the test compounds exhibited better minimum inhibitory concentration (MIC) against the test isolates. The MIC of the compounds against Staphylococcus species and E. coli was discovered to be higher when compared to Klebsiella species and Salmonella species. The piperidine compounds that were used as alternatives showed promising susceptibility towards pathogens.
Healthcare-associated infections, Nosocomial pathogens, Piperidines.
Nosocomial infections are the healthcare-associated infections that are seen among patients globally, both in underdeveloped and developed countries1. While in hospital, patients get exposed to pathogens through various sources like healthcare staff, environment, and also from other infected patients2. Among which low resistance of the patients getting contact with drug resistance of endemic organisms was the leading cause of death and increased morbidity in hospitalized patients3. A large number of microorganisms are responsible for hospital infection. They can be natural flora of the patient and cause infection or multidrug resistance (MDR) pathogen obtained from the hospital environment or through staff-patient contact4,5.
The most prevalent drug-resistant organisms that are met in the health care settings are methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum beta-lactamase (ESBL)- producing Enterobacteriaceae, multidrug-resistant Pseudomonas aeruginosa, Enterococcus faecium, Klebsiella pneumoniae, and Acinetobacter baumannii6-9. These pathogens are commonly known to be ESKAPE pathogens, and these are majorly accountable for nosocomial infections and are skilled in “escaping” the antimicrobial agent’s action10,11. These pathogens possess the ability to irreversible cleavage catalyzed by modifying enzyme of the target site of antibiotic, reduced permeability or increased efflux of the drug to tackle the action of antibiotic against it12. World Health Organization (WHO) has also recorded ESKAPE pathogens in the list of 12 bacteria for which there is a very urgent requirement of new antibiotics13.
Antibiotic resistance among these pathogens due to constant exposure of antibiotic prophylaxis plays a critical role in treating these pathogens14,15. Chaudhary16 reported a comparison reports available in World Health Organization’s (WHO), European Union, the Centers for Disease Control and Prevention (CDC) and Global Antibiotic Resistance Partnership (GARP) have estimated around 23,000 in the US, 25,000 in EU, and 58,000 in India as per annum death toll from numerous infections due to drug-resistant bacteria. Nearly 70 % of bacterial infections are resistant to one or several antibiotics, which are used regularly to eradicate their disease17.
At present alternative therapies are used or under various stages of the trail are drugs that are in combination or use of antibacterial antibodies, antimicrobial peptides, nanoparticles, photodynamic therapy, and phytochemicals as antibacterial agents18,19. Numerous alternative therapies are showing promising results in-vitro, but the same is not available in-vivo due to one or many limitations20. In this context, piperidine compounds can also be used as a potential alternative therapeutical compound. Piperidine scaffold has shown a decisive role in several pharmaceutical drugs that are presently existing21,22. Piperidine derivatives are isolated from plants and are also synthesized using one or more of their various chemical reactions that have been recognized for the synthesis of piperidine derivatives22,23. Piperidine compounds not only significantly reducing bacterial invasion but also exhibit an attenuated pro-inflammatory response in host-bacteria interactions24. There is numerous historical background which supports the effectiveness of piperidine derivatives as a promising antimicrobial agent and exhibits significant minimum inhibitory concentration when compared to standard drugs21-24. Hence, the present study focused on isolating multi-drug nosocomial pathogen and testing the efficiency of piperidine derivatives against them.
Sample processing
During the study period from November 2018 to March 2019, a total of 354 clinical swabs samples such as aspirates and pus samples of abscesses, surgical, accidental wound infections, and urinary catheters along with urine samples were collected from inpatients of Intensive care facilities, surgical and post-operative wards in multi-specialty hospitals in and around Tamil Nadu, India. Samples were collected from patients who had no history of prior infection. Samples were collected from the surgical wound of patients after three days of surgery in a hospital with the help of Staff Nurses by obtaining prior permission from the hospital and patient. The samples were collected aseptically and immediately transferred to the microbiology laboratory for further study.
Identification of isolates
The swab of aspirates, pus, and catheters was serially diluted and inoculated in Luria Bertani (LB) broth and incubated at 37°C. After incubation for about 24 h, one loopful of the culture was streaked on selective agar medium. The selective agar medium that was used for the isolation of enteric pathogens is listed in table 1. Further, the isolates were identified by Gram’s staining and series of biochemical tests.
Table (1):
Selective medium for isolation of pathogens.
Selective media |
Bacterial genera |
|---|---|
Mannitol Salt Agar |
Staphylococcus sp. |
Xylose Lysine Deoxycholate Agar |
Salmonella sp. |
Eosine Methylene Blue agar (EMB) |
E. coli |
Mac Conkey Agar |
Klebsiella sp. |
Cetrimide Agar |
Pseudomonas sp. |
Antimicrobial susceptibility testing
The Kirby-Bauer disc diffusion method using Mueller-Hinton agar (MHA) was used for testing the susceptibility of the isolates. As suggested by the Clinical and Laboratory Standards Institute (CLSI)25 antibiotic disc strengths were used where CLSI’s criteria for testing susceptibility and resistance were shadowed. About ten antibiotics were used to assess the antimicrobial sensitivity pattern of the isolates, and the medicines which were prescribed on a routine base to the patients were utilized. Zones of inhibition around the antibiotic disc were measured using a normal measuring scale and noted.
Quantitative antibacterial activity assay of piperidine compounds
Minimum inhibitory concentration (MIC) of piperidine compounds against the isolated pathogens was determined by measuring the OD value at 600 nm, and all tests were performed in duplicates. DMSO was used as a solvent blank. RPMBD (2-methyl-2,4-diphenylbenzodiazepine), CARPMBD (1-chloroacetyl-2-methyl-2,4-diphenylbenzodiazepine), PM3MPO (3-Methyl-2,6-diphenyllpiperidin-4-one), PCPM3MPO (1-phenylcarbamoyl-3-methyl-2,6-diphenyllpiperidin-4-one), NPM3MPO (1-nitroso-3-methyl-2,6-diphenyllpiperidin-4-one), PM3EPO (3-ethyl-2,6-diphenyllpiperidin-4-one), MPM3EPO (1-methoxycarbonyl-3-ethyl-2,6-diphenyllpiperidin-4-one) are the piperidine compounds used in this study. DMSO and piperidine compounds in the varying concentration ranging from 6.25, 12.5, 25, 50, 100, and 200µg/mL were added in 96-well microtitre plate. Tetracycline, penicillin, and vancomycin were used as a reference for comparing the susceptibility pattern exhibited by test isolates. Bacterial cultures grown overnight were adjusted to 0.5 McFarland standard and from that 100µL were added to each well. The positive control titre well was added with sterile broth without any test compound. Micro dilution plates sealed with a tight lid before incubation in order to prevent desiccation and contamination. The plates were incubated at 37°C for 24 h and MIC was read.
Isolation and identification of pathogens
About 282 swab samples from wounds, pus, abscesses, urinary catheters, and 72 urine samples were collected aseptically and transferred in the microbiology laboratory for further processing. The distribution of samples from various clinical specimens are listed in Table 2, and the biochemical profile of positive isolates are listed in Table 3. The organism and the source from which it was been isolated are tabulated in Table 4.
Table (2):
Distribution of samples among various clinical specimens.
S. No |
Types of sample |
No. of samples Collected |
No. of positive sample |
No. of negative sample |
|---|---|---|---|---|
1. |
Pus |
89 |
43 |
46 |
2. |
Pus Aspirate |
64 |
30 |
34 |
3. |
Urine |
72 |
43 |
29 |
4. |
Urinary Catheter |
68 |
26 |
42 |
5. |
Abscesses |
61 |
18 |
43 |
Total |
354 |
160 |
194 |
Table (3):
Typical biochemical profile of isolates.
Gram’s staining |
Indole |
MR |
VP |
Urease |
TSI |
Catalase |
Glucose |
Lactose |
Maltose |
Sucrose |
Motility |
Oxidase |
Suspected organism |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
G+ve Cocci |
– |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
– |
– |
Staphylococcus sp. |
G-ve Rods |
– |
+ |
– |
– |
+ |
+ |
+ |
– |
+ |
– |
+ |
– |
Salmonella sp. |
G-ve Rods |
– |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
+ |
+ |
Pseudomonas sp. |
G-ve Rod |
– |
+ |
– |
+ |
+ |
+ |
+ |
– |
– |
+ |
+ |
– |
Proteus sp. |
G-ve Rod |
– |
– |
+ |
– |
+ |
+ |
– |
+ |
– |
– |
+ |
– |
E. coli |
G-ve Rod |
– |
+ |
– |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
– |
– |
Klebsiella sp. |
(-) Negative(+) Positive
Table (4):
Identification of isolates.
S. No |
Isolate |
No. of Positives |
Source |
|---|---|---|---|
1. |
Staphylococcus sp. |
37 |
Pus (15), Aspirate (11), Urine (8), Abscesses (3) |
2. |
Salmonella sp. |
23 |
Aspirate (4), Urine (14), Catheter (5) |
3. |
Pseudomonas sp. |
27 |
Pus (11), Aspirate (4), Urine (12) |
4. |
Proteus sp. |
21 |
Pus (3), Aspirate (5), Urine (13) |
5. |
E. coli |
34 |
Pus (7), Urine (16), Aspirate (2), Catheter (9) |
6. |
Klebsiella sp. |
18 |
Pus (1), Urine (10), Catheter (7) |
Antibiogram of the isolates
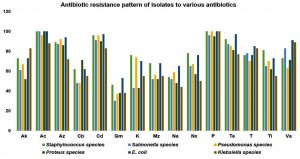
All the 160 isolates obtained were screened for the antibiotic sensitivity against 15 commercially available antibiotics. About 98% of resistance was noticed against amoxicillin and penicillin, 93% towards cefdinir, 88% against azithromycin and tetracycline, 78% against teicoplanin and vancomycin, 72% towards amikacin, 70% towards ticarcillin 68% norfloxacin, 63% against kanamycin, 60%, 58%, 55% resistance towards mezlocillin, carbenicillin, nalidixic acid and 42% against gentamycin respectively. E. coli and Staphylococcus species isolates exhibited the highest degree of resistance among the isolates, and results are presented as figure 1. Among the positive isolates tested Staphylococcus species showed multi-drug resistance.
Minimum inhibitory concentration of piperidine compounds
One isolate from each organism, which shows the highest drug resistance was selected for studying the MIC of piperidine compounds, and results are tabulated in Table 5. NPM3MPO compound was found to be effective against Staphylococcus species at the lowest concentration (50µg/mL), followed by PM3EPO and MPM3EPO at 100µg/mL. PM3MPO exhibited the least inhibition. The entire test compound showed inhibition of Salmonella species at 12.5µg/mL except for PM3EPO and PCPM3MPO. PM3EPO shows inhibition at 25µg/mL but PCPM3MPO couldn’t prove its inhibition at test concentration.
Table (5):
MIC of piperidines against test isolates at 600 nm.
S.No |
OD Value at 600 nm for test compounds |
Staphylococcus sp. (µg/mL) |
Salmonella sp. (µg/mL) |
Pseudomonas sp. (µg/mL) |
Proteus sp. (µg/mL) |
E. coli (µg/mL) |
Klebsiella sp. (µg/mL) |
|---|---|---|---|---|---|---|---|
1 |
Tetracycline |
100 |
25 |
50 |
50 |
100 |
12.5 |
2 |
Penicillin |
100 |
50 |
100 |
50 |
200 |
25 |
3 |
Vancomycin |
100 |
50 |
50 |
50 |
100 |
25 |
4 |
RPMBD |
50 |
12.5 |
100 |
50 |
– |
6.25 |
5 |
CARPMBD |
– |
25 |
– |
200 |
200 |
– |
6 |
PM3MPO |
– |
12.5 |
100 |
100 |
– |
6.25 |
7 |
PCPM3MPO |
200 |
– |
100 |
50 |
– |
– |
8 |
NPM3MPO |
50 |
12.5 |
100 |
50 |
– |
6.25 |
9 |
PM3EPO |
100 |
25 |
100 |
200 |
– |
– |
10 |
MPM3EPO |
100 |
12.5 |
– |
50 |
100 |
6.25 |
The majority of test compounds showed inhibition of Pseudomonas species at 100µg/mL. Most of the test isolates inhibit Proteus species at 50µg/mL concentration, whereas CARPMBD showed the least restraint against test isolate. Against E. coli, MPM3EPO inhibits test isolate at 100µg/mL and PM3MPO at 200µg/mL. Salmonella species, Klebsiella species showed the least resistance towards tested compounds. Most of the compound inhibits the test isolates at 6.25µg/mL and 12.5µg/mL. Similar to the results of antibiotic resistance study in Fig. 1, E. coli and Staphylococcus species showed the highest resistance towards test compounds when compared to other test isolates.
In nosocomial infections, antibiotic-resistant microorganisms likely lead to less than ideal clinical outcomes when related to those that are showing susceptibility to antibiotics. The frequent occurrence of antibiotic resistance has presented extraordinary challenges for researchers and clinicians. Alternative treatments or fresh new medicines should be developed to tackle this challenge. This study is focused on developing piperidine derivatives as alternative therapeutics for treating multidrug-resistant pathogens.
The samples for isolation of pathogens from various nosocomial infections were obtained from surgical and post-operative wards after 48 h of admission. Ginawi et al.1 also isolated the pathogens after 48 h of admission in the ward. They reported that among nosocomial infection, 27.2% Gram-positive, and 72.8% were Gram-negative. Similarly, in the present study, 23% of isolates were Gram-positive and 77% were Gram-negative. This result also concords with Vanitha et al.26 and Dutta and Gupta27 reports, Gram-negative bacilli have taken over the Gram-positive organisms, especially in hospital settings. Feleke et al.28 obtained a high rate of Gram-negative bacteria as the causative agents of nosocomial infections than Gram-positives.
In the present study, the isolation rate of pathogens recorded as Staphylococcus species (23%), followed by E. coli (22%), Pseudomonas species (17%), Salmonella species (14%), Proteus species (13%) and Klebsiella species (11%). In their study, Ginawi et al.1 recorded the isolation rate of P. aeruginosa (43.5%) and S. aureus (34.8%). Like the present study, Sohrabi et al.29 obtained E. coli (64.3%), coagulase-negative Staphylococci (11.2%), and Klebsiella species (8.1%) as common pathogens. This agrees with Nwadioha et al.30 report that E. coli (51.0%), Klebsiella species (19.6%) and Proteus mirabilis (10.0%) were the common isolates. Mulu et al.31 in his research, obtained S. aureus (100) as the predominant isolate, followed by E. coli (39), P. aeruginosa (30), Salmonella species (25) and Klebsiella species (7) which is similar to that of this present study.
While screening for antibiotic resistance, about 98% of test isolates in present study were resistance towards amoxicillin and penicillin, cefdinir (93%), azithromycin and tetracycline (88%), teicoplanin and vancomycin (78%), amikacin (72%), towards ticarcillin (70%), norfloxacin (68%), 63% against kanamycin, 60%, 58%, 55% resistance towards mezlocillin, carbenicillin, nalidixic acid and 42% against gentamycin respectively. E. coli and Staphylococcus species isolates exhibited the highest degree of resistance among the strains.
About 46% of Staphylococcus species, 30% of Salmonella species, Pseudomonas species (37%), Proteus species (38%), E. coli (53%) and 38 % of Klebsiella species showed resistance against gentamycin and it stands as low resisting antibiotic according to the present study. In the case of amikacin, Klebsiella species exhibited the highest degree of resistance. E. coli registered its highest potential to resist azithromycin (94%), cefdinir (97%), gentamycin (53%), nalidixic acid (65%), tetracycline (97%), teicoplanin (85%) and vancomycin (91%). Like, Staphylococcus species also showed predominant resistance against kanamycin (76%), norfloxacin (78%) and ticarcillin (81%). E. coli showed the highest resistance than Staphylococcus species, Proteus species and Salmonella species exhibited the lowest degree of resistance among the isolated organisms.
Gashe et al.32 assessed the antimicrobial susceptibility pattern of clinical isolates, which shows most isolates are revealing a higher resistance rate (>80%) to amoxicillin, amoxicillin/clavulanic acid, ampicillin, chloramphenicol, and cloxacillin. Gram-positive bacteria show increased resistance levels to ampicillin, cloxacillin (92.4), chloramphenicol, and amoxicillin (84.8-97.4%). E. coli has shown an increased level of resistance against ampicillin, 49 (100%), amoxicillin, 42 (85.7), chloramphenicol, 41 (83.7%), and tetracycline 40 (81.6%). Even in the present study, E. coli and Staphylococcus species. isolates exhibited the highest degree of resistance among the isolates towards various antibiotics.
Among the test compounds, PM3MPO and MPM3EPO showed the least inhibition against the isolates. Towards PCPM3MPO, Staphylococcus species and Pseudomonas species showed resistance, and E. coli showed complete resistance towards CARPMBD. In the study conducted by Letafat et al.33 piperazinyl quinolones high inhibition against methicillin-resistant S. aureus, E. coli ATCC 8739, K. pneumoniae ATCC 10031 and P. aeruginosa ATCC 9027 were comparable or higher concerning the reference drugs ciprofloxacin and norfloxacin. Similarly, in the present study, the majority of the test compounds exhibit higher inhibition when compared with reference drugs used in this study. Similarly, Desai et al.34 conducted the antimicrobial activity of thiazole derivatives by using Ciprofloxacin as reference compounds. Thiazole compounds show inhibition activity 25µg/mL to 50µg/mL against E. coli, MIC 50µg/mL against P. aeruginosa and MIC of 50µg/mL against S. aureus respectively. But in the present study, the piperidine compounds showed the least inhibition against thiazole compounds.
In the study conducted by Imran et al.35 the MIC of test compounds ranged between 25- 50µg/mL against E. coli, P. aeruginosa, K. pneumonia, S. aureus, and P. vulgaris. But in the case of the present study, the MIC range varies between 6.25 to 200µg/mL. Salmonella species and Klebsiella species showed the least resistance towards tested compounds. Most of the compound inhibits the test isolates at 6.25µg/mL and 12.5µg/mL. The MIC of E. coli and Staphylococcus species were high as like antibiotic resistance when compared to other test isolates which range above 100µg/mL.
Arslan et al.36 studied the antimicrobial activity of some piperidine and pyrrolidine highest antimicrobial activities with MICs of 32-128 mg/mL, but the low activity of the compounds was noted against E. coli and K. pneumoniae, that are highly pathogenic Gram-negative bacteria to humans. Indeed, K. pneumoniae had shown lower microbial susceptibility when compared with S. aureus and C. albicans. Kaur et al.37 reported that all the synthesized compounds showed better antimicrobial activity at MIC 50µg/mL. Similarly, the compound synthesized by Naicker et al.23 showed MIC of 1.5 mg/mL against E. coli, P. aeruginosa, S. aureus, B. cereus and B. subtilus. Agreeing results were obtained in the study conducted by Duruskari et al.38. The synthesized compounds possess pronounced antimicrobial activity against pathogenic bacteria Acinetobacter baumannii BDU32, E. coli BDU12, K. pneumoniae BDU44, P. aeruginosa BDU49, and S. aureus BDU23 in well diffusion method.
Microbial evolution is not a reversible process. As for which approach to combating antibiotic resistance will ultimately prevail, that is still up for debate. In this study, search for new therapeutics in treating pathogens was carried out, where piperidine compounds were used as alternatives to study the pattern of antibiotic susceptibility over the positive isolates that were isolated from nosocomial patients. Overall, the piperidine compounds exhibited better results than that of regular antibiotics that are in general use. These results show the importance of further research in this field of alternative antibiotics to combat emerging deadly pathogens.
ACKNOWLEDGMENTS
Not applicable.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS’ CONTRIBUTION
We declare that this work was done by the authors named in this article and all liabilities pertaining to claims relating to the content of this article will be borne by the authors. Both authors R V and T V have equally contributed to this study in designed this study, carrying out laboratory works, collected and analysed the data, and also prepared the manuscript.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All data generated or analysed during this study are included in this published article. If any specific data required, then available on request from the authors.
- Ginawi I, Salee MM, Sigh M, Vaish K, Ahmad I, Srivastava VK, Abdullah FM. Hospital acquired infections among patients admitted in the medical and surgical wards
of a non-teaching secondary care hospital in Northern India. J Clin Diag Res, 2014; 8: 81-83. - Ducel JF, Nicolle L. Prevention of hospital-acquired infections. Geneva: WHO, 2002.
- Apostolopoulu E, Katsaris G. Socioeconomic impact of Nonsocomial infections. Icus Nurs Web J, 2003.
- Suresh G, Joshi GML. Acinetobacter baumannii: an emerging pathogenic threat to public health. World J Clin Infect Dis, 2013; 3: 25-36.
Crossref - Jayanthi A. Most common healthcare-associated infections: 25 bacteria, viruses causing HAIs, Becker’s hospital review. 2014.
- Jimenez-Alcaide E, Medina-Polo J, Garcia-Gonzalez L. Healthcare-associated urinary tract infections in patients with a urinary catheter: Risk factors, microbiological characteristics and patterns of antibiotic resistance. Arch Esp Urol, 2015; 68: 541-550.
- Nadimpalli M, Stewart JR, Pierce E. Livestock associated, antibiotic-resistant Staphylococcus aureus nasal carriage and recent skin and soft tissue infection among
industrial hog operation workers. PLoS One, 2016; 11: 165713. - Apostolopoulu E, Stergiopoulou A, Telalidou K, Konstantopoulou G, Giannatou M, Skotis I. Socioeconomic impact of nosocomial infections in surgical inntensive care unit. Icus Nurs Web J, 2005.
- Dantas RC, Ferreira ML and Gontijo-Filho PP. Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol, 2014; 63:1679-1687.
Crossref - Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis, 2008; 197: 1079-1081.
Crossref - Navidinia M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. J Paramed Sci, 2016; 7: 4978.
- Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int, 2016; 2475067.
Crossref - Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL. Discovery, research and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis, 2018; 18: 318-327.
Crossref - Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol, 2007; 5: 48-56.
Crossref - Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health, 2017; 10: 369-378.
Crossref - Chaudhary AS. A review of global initiatives to fight antibiotic resistance and recent antibiotics’ discovery. Acta Pharm Sin B, 2016; 6: 552-556.
Crossref - Duruskari GSH, Maharramov MN, Hasanova USH, Ganbarov KHG, Eyvazova GM, Israyilova AA, Maharramov AM. Synthesis and Antimicrobial Properties of New Derivatives of Morpholine and Piperidine Based on 1-Chloro-3-methoxy-propylbenzene. Asian J Chem, 2018, 30: 269-272.
Crossref - Mandal SM, Roy A, Ghosh AK, Hazra TK, Basak A, Franco OL. Challenges and future prospects of antibiotic therapy: from peptides to phages utilization. Front Pharmacol, 2014; 5: 105.
Crossref - Kaur I. Novel strategies to combat antimicrobial resistance. J Infect Dis Ther, 2016; 4: 292.
Crossref - Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front Microbiology, 2019; 10: 539.
Crossref - Das S, Brahmachari G. Ni(ClO4)2.6H2O: An efficient catalyst for one-pot synthesis of densely functionalized piperidine scaffolds via multicomponent reaction in ethanol at room temperature. J Organic Biomolecular Chem, 2013; 1: 33-46.
- Anthal S, Brahmachari G, Das S, Kant R and Gupta VK. Ethyl4-anilino-2, 6-bis (4-fluorophenyl)-1-phenyl-1,2,5,6-tetrahydropyridine-3- carboxylate. Acta Cryst, 2013; 69: 299-300.
Crossref - Naicker L, Venugopala, Narayanaswamy K, Shode F, Odhav B. Antimicrobial and antioxidant activities of piperidine derivatives. Afr J Pharm Pharmacol, 2015; 9: 783-792.
Crossref - Kohler H, Rodrigues SP, Maurelli AT, McCormick BA. Inhibition of Salmonella typhimurium enteropathogenicity by piperidine, a metabolite of the polyamine cadaverine. J Infect Diseases, 2002; 186: 1122-1130.
Crossref - Clinical and Laboratory Standards Institute., Performance standards for antimicrobial susceptibility testing: eighteenth informational supplement: approved standards M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA; 2008.
- Vanitha RN, Gopal K, Narendra VM, Vishwakanth D, Nagesh VRD, Yogitha M. A retrospective study on blood stream infections and antibiotic susceptibility patterns in a tertiary care teaching hospital. Int J Pharm Sci, 2012; 4: 543-548.
- Dutta MMP, Gupta V. Antimicrobial susceptibility pattern of blood isolates from a teaching hospital in North India. Jpn J Infect Dis, 2005;
58: 174-176. - Feleke T, Eshetie S, Dagnew M, Endris M, Abebe W, Tiruneh M, Moges F. Multidrug-resistant bacterial isolates from patients suspected of nosocomial infections at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Res Notes, 2018; 11: 602.
Crossref - Sohrabi MB, Khosravi A, Zolfaghari P, Sarrafha J. Evaluation of nosocomial infections in Imam Hossein (as) Hospital of Shahrood, 2005. Journal of Birjand University of Medical Sciences, 2009; 16: 33-39.
- Nwadioha SI, Nwokedi EE, Jombo GTA, Kashibu E, Alao OO. Antibiotics susceptibility pattern of uropathogenic bacterial isolates from community and hospital acquired
urinary tract infections in a Nigerian tertiary hospital. The Internet J Infect Dis, 2010; 8. - Mulu W, Abera B, Yimer M, Hailu T, Ayele H, Abate D. Bacterial agents and antibiotic resistance profles of infections from diferent sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia: a cross-sectional study. BMC Res Notes, 2017; 10: 254.
Crossref - Gashe F, Mulisa E, Mekonnen M, Zeleke G. Antimicrobial resistance profile of different clinical isolates against third-generation Cephalosporins. J Pharmaceutics, 2018; 5070742.
Crossref - Letafat B, Emami S, Mohammadhosseini N, Faramarzi MA, Samadi N, Shafiee A, Foroumadi A. Synthesis and antibacterial activity of new N-[2-(Thiophen-3-yl)ethyl]
piperazinyl quinolones. Chem Pharm Bull, 2007; 55: 894-898.
Crossref - Desai NC, Makwana AH, Rajpara KM. Synthesis and study of 1,3,5-triaine based thiazole derivatives as antimicrobial agents. J Saudi Chem Society, 2016; 20: 334-341.
Crossref - Imran M, Alam O, Abida A. Synthesis and antimicrobial activity of some 2-Piperidinomethylamino-4-(7-H/substituted coumarin-3-yl)-6-chlorosubstitutedphenyl pyrimidines. Trop J Pharm Res. 2016; 15: 1965.
Crossref - Arslan S, Logoglu E, Oktemer A. Antimicrobial activity studies on some piperidine and pyrrolidine substituted halogenobenzene derivatives. J Enzyme Inhib Med Chem, 2006; 21: 211-214.
Crossref - Kaur G, Balamurugan P, Vasudevan S, Jadav S, Princy SA. Antimicrobial and Antibiofilm Potential of Acyclic Amines and Diamines against Multi-Drug Resistant Staphylococcus aureus. Frontiers in Microbiology, 2017; 8: 1767.
Crossref - Duruskari, Maharramov MN, Hasanova USH, Ganbarov KHG, Eyvazova GM, Israyilova AA, Maharramov AM. Synthesis and Antimicrobial Properties of New Derivatives of Morpholine and Piperidine Based on 1-Chloro-3-methoxy-propylbenzene. Asian J Chem, 2018; 30: 269-272.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.