ISSN: 0973-7510
E-ISSN: 2581-690X
Multidrug-resistant (MDR) microorganisms that cause septicemia are responsible for high morbidity and mortality rates among patients in extreme age groups and pose serious treatment challenges despite multipronged measures. This hospital-based study was conducted in the Postgraduate Department of Microbiology at a Tertiary Care Hospital of Kashmir (India) to investigate the prevalence and antimicrobial resistance patterns of bacteria that cause neonatal sepsis. As the resistance pattern of causative agents varies among hospitals, the study results suggest an appropriate empirical therapy for neonatal sepsis needs to be devised in our hospital. Neonatal sepsis, diagnosed by clinicians in the neonatal intensive care unit on the basis of clinical and laboratory findings, was categorized as early-onset sepsis (EOS) or late-onset sepsis (LOS) according to whether clinical presentation occurred at <72 h after birth or between 72 h and 28 days of life, respectively. Blood samples collected from 1200 neonates were cultured in a BacT/ALERT® 3D system (bioMerieux, Inc., Durham, NC, USA). Bacterial identification and antimicrobial susceptibility tests were performed using a VITEK 2 Compact system (bioMerieux). Of the 1200 blood cultures, 126 (10.5%) were bacteremia positive. Of these 126 cultures, 73 (58%) contained gram-positive bacteria, which occurred predominantly in the EOS group (p < 0.001), whereas 53 (42%) contained gram-negative bacteria, which were equally distributed in both groups. Bacteremia was more common in the EOS group (n = 88; 70%), with Staphylococcus aureus (n = 25; 20%) being the predominant isolate, followed by Klebsiella pneumoniae (n = 20; 15.9%). In total, 38 (30%) isolates were recovered in the LOS group, with Klebsiella pneumoniae (n = 11; 9%) being predominant, followed by Acinetobacter baumannii (n = 9; 7%). Fifty-six (44.4%) and 28 (22.2%) isolates from the EOS and LOS groups, respectively, were MDR, with Staphylococcus spp. (n = 49; 38.9%), Klebsiella pneumoniae (n = 17; 13.5%), and Acinetobacter baumannii (n = 14; 11.1%) in particular showing high resistance rates. With the high prevalence of MDR infections, colistin and vancomycin can be used as initial empirical therapies in our hospital setting. This study underscores the urgent need for robust antimicrobial stewardship and infection control measures to combat the increasing threat of MDR bacterial infections in neonates.
Neonatal Sepsis, Blood Culture, Antimicrobial Susceptibility Profile, Multidrug-resistance
In low- and middle-income countries, sepsis is the main cause of morbidity among neonates and responsible for 99% of the global burden of neonatal deaths. Without a significant decrease in infection-related neonatal deaths, it would be impossible to achieve Sustainable Development Goal 3, which aims to decrease the neonatal mortality rate to at least 12 per 1000 live births by 2030.1 When sepsis is suspected in neonates, antimicrobials are administered empirically to prevent severe consequences, as the early diagnosis of neonatal septicemia is difficult.2 However, the use of broad-spectrum antimicrobial agents in empirical therapy inadvertently leads to the development of multidrug-resistance among the causative agents of sepsis, resulting in a high economic burden. Organisms that acquire resistance to at least one antimicrobial agent in three or more antimicrobial classes are defined as multidrug-resistant (MDR).3 Given that antimicrobial resistance is a major public health problem, blood culture and antimicrobial sensitivity testing must be performed in all patients with suspected sepsis before initiating antimicrobials, as bacterial isolation from blood culture remains the gold standard for diagnosing bacteremia.4 Early recognition of the disease and the prompt administration of appropriate antimicrobials can improve the outcomes of afflicted neonates.2 However, because the microbial spectrum of bacteremia and the antibiogram of the causative pathogens show interregional variation and even vary among hospitals in the same region, a single antibiotic regimen cannot be recommended in all settings. Hence, once initiated, antimicrobials should be modified according to antimicrobial susceptibility reports. Antibiotic stewardship, defined as the set of practices followed to ensure antibiotics are chosen and administered safely and appropriately, should be adopted by all healthcare settings. In this era of antibiotic-resistance, the changing microbiological patterns of bacteremia warrant an assessment of the causative agents and their patterns of susceptibility to antimicrobials. Hence, this study was conducted to identify the antimicrobial resistance patterns of bacteria isolated from neonates presenting with sepsis within 72 h after birth (early-onset sepsis; hereinafter EOS) or between 72 h and 28 days of age (late-onset sepsis; hereinafter LOS).5,6
This prospective hospital-based study was conducted in the Postgraduate Department of Microbiology at a Tertiary Care Hospital of Kashmir (India). This hospital is equipped with a well-functioning level-III neonatal intensive care unit (ICU) where sick neonates (from birth to 28 days of life) are admitted. In total, 1200 neonates with a provisional clinical diagnosis of sepsis were admitted in the neonatal ICU during the 18-month study period, accounting for 44% of total admissions in the unit (N = 2731). After obtaining due approval by the Institutional Ethics Committee (SIMS1131/IEC/-SKIMS/2018-244), blood samples were collected aseptically from the 1200 neonates into BacT/ALERT® PF Plus (bioMerieux, Inc., Durham, NC, USA) aerobic pediatric blood culture bottles. Neonatal sepsis was diagnosed by clinicians in the neonatal ICU on the basis of clinical signs of sepsis, such as fever, lethargy, and respiratory distress, along with specific laboratory criteria, such as abnormal white blood cell counts and C-reactive protein levels.
Isolates recovered from neonates who presented with symptoms of sepsis within 72 h of birth were grouped as causative agents of EOS, whereas those recovered from patients between 72 h and 28 days of age were grouped as pathogens causing LOS.5
Inclusion criteria
Bacterial isolates from flagged-positive bottles of blood cultures from neonates. Flagged-positive blood culture bottles are those in which the positive growth of organisms had been detected using the BacT/ALERT® 3D Automated Microbial Detection System (bioMerieux).
Exclusion criteria
Flagged-negative blood culture bottles from neonates with a probable diagnosis of sepsis and isolates other than bacteria obtained from flagged-positive blood culture bottles.
Identification and antimicrobial susceptibility testing of study isolates
Under aseptic measures, 1-5 ml of venous blood (4-4.5% of total blood volume) was drawn directly into BacT/ALERT® PF Plus (bioMerieux) aerobic pediatric blood culture bottles. Then, the bottles were checked for labeling errors regarding patient details before being loaded onto the BacT/ALERT® 3D Microbial Detection System. While maintaining aseptic measures, a 3 ml syringe was used to dispense a drop of blood from the flagged-positive blood culture bottle onto a slide for Gram staining. On the basis of the staining results, a sample from the bottle was inoculated onto selective and differential media and incubated at 37 °C for 18-24 h. Subsequently, growth was inspected, and Gram staining of the culture was performed again to confirm the findings. Then, inocula were prepared for processing on a VITEK 2 compact system (bioMerieux) to identify the organisms to the species level and for antimicrobial susceptibility testing as per the Clinical Laboratory Standards Institute guidelines.7 Aside from gram-positive methicillin-resistant Staphylococcus spp., gram-negative isolates resistant to at least one antimicrobial agent from each of the three antimicrobial classes were defined as MDR.3
Statistical analysis
Data analysis was performed using SPSS v.25.0 software. Continuous variables were calculated using Student’s t-test. For the 2-tailed test results, a p–value of less than 0.05 was considered statistically significant.
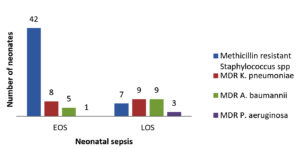
The 1200 neonates with a provisional diagnosis of sepsis made up 44% of the total admissions in the neonatal ICU during the study period (N = 2731). Among them, 126 (10.5%) had microbiologically confirmed bacteremia (Table 1). The representation of both sexes was comparable, with a male-to-female ratio of 1:0.7. As shown in Table 2, the gram-positive isolates (n = 73, 58%) comprised Staphylococcus spp. (n = 68) [methicillin-resistant Staphylococcus aureus (MRSA) (n = 20); methicillin-sensitive Staphylococcus aureus (n = 7); methicillin-resistant Staphylococcus epidermidis (n = 16); methicillin-sensitive Staphylococcus epidermidis (n = 2); methicillin-resistant Staphylococcus haemolyticus (n = 8); methicillin-sensitive Staphylococcus haemolyticus (n = 6); methicillin-resistant Staphylococcus hominis, (n = 5); and methicillin-sensitive Staphylococcus hominis (n = 4) and Enterococcus spp. (n = 5). All 68 Staphylococcus isolates were resistant to penicillin, whereas 80% (n = 54) were resistant to erythromycin, 65% (n = 44) to clindamycin, and 50% (n = 34) to trimethoprim/sulfamethoxazole. Among the MDR isolates (n = 84), 58.3% (n = 49) were methicillin-resistant Staphylococcus spp. (Figure), with MRSA alone accounting for 23.8% (n = 20). Of the Enterococcus faecium isolates, 80% (n = 4) were resistant to penicillin and 60% (n = 3) to ampicillin (Table 3). However, none of the gram-positive isolates showed resistance to linezolid, teicoplanin, or vancomycin.
Table (1):
Incidence of bacteremia in Neonatal sepsis
| Frequency (N) | Percentage (%) | ||
|---|---|---|---|
| Probable sepsis (Total Samples Received) | 1200 | 100% | |
| Culture positive samples with significant growth (n = 178, 14.8%) | Bacteremia | 126 | 10.5% |
| Candidemia | 52 | 4.3% | |
| Sterile samples | 1022 | 85.2% | |
Table (2):
Association of various bacterial isolates with neonatal sepsis
| Bacterial isolates (N = 126) | EOS | LOS | Total Frequency N | Total % | |
|---|---|---|---|---|---|
| Frequency N (%) | Frequency N (%) | ||||
| Gram-positive Isolates | 63 (50.0) | 10 (8.0) | 73 | 58.0 | |
| Staphylococcus aureus | MR | 18 (14.3) | 2 (1.6) | 20 | 15.9 |
| MS | 07 (5.6) | 0 | 7 | 5.5 | |
| Staphylococcus epidermidis | MR | 13 (10.3) | 3 (2.4) | 16 | 12.7 |
| MS | 02 (1.6) | 0 | 2 | 1.6 | |
| Staphylococcus haemolyticus | MR | 07 (5.6) | 1 (0.8) | 8 | 6.3 |
| MS | 06 (4.8) | 0 | 6 | 4.8 | |
| Staphylococcus hominis | MR | 04 (3.2) | 1 (0.8) | 5 | 4.0 |
| MS | 04 (3.2) | 0 | 4 | 3.2 | |
| Enterococcus faecium | 02 (1.6) | 3 (2.4) | 5 | 4.0 | |
| Gram-negative Isolates | 25 (19.8) | 28 (22.0) | 53 | 42.0 | |
| Klebsiella pneumoniae | 09 (7.0) | 11 (8.6) | 20 | 15.8 | |
| Acinetobacter baumannii | 05 (4.0) | 9 (7.0) | 14 | 11.1 | |
| Pseudomonas aeruginosa | 02 (1.6) | 4 (3.1) | 6 | 4.7 | |
| Escherichia coli | 05 (4.0) | 0 | 5 | 4.0 | |
| Burkholderia cepacia complex | 01 (0.8) | 1 (0.8) | 2 | 1.6 | |
| Enterobacter cloacae | 3 (2.4) | 1 (0.8) | 4 | 3.2 | |
| Serratia spp | 0 | 2 (1.6) | 2 | 1.6 | |
| TOTAL | 88 (70) | 38 (30) | 126 | 100 | |
MR: Methicillin-resistant; MS: Methicillin Sensitive
Table (3):
Antibiotic susceptibility profile of Gram-positive isolates recovered from cases of neonatal sepsis
| Antibiotics | Antibiotic susceptibility profile of Staphylococcus spp. (N = 68) | Antibiotic susceptibility profile of Enterococcus spp. (N = 5) | ||
|---|---|---|---|---|
| Sensitive N (%) | Resistant N (%) | Sensitive N (%) | Resistant N (%) | |
| Cefoxitin | 19 (28) | 49 (72 ) | * | * |
| Ampicillin (Amoxicillin,Ampicillin sulbactum,Amoxicillin/ clavulonic acid, Piperacillin/ tazobactum) | * | * | 2(40) | 3(60) |
| Penicillin | 0 | 68 (100) | 1 (20) | 4 (80) |
| Erythromycin | 14 (20) | 54 (80) | IR | IR |
| Clindamycin | 24 (35) | 44 (65) | * | * |
| Cotrimoxazole | 34 (50) | 34 (50) | * | * |
| Vancomycin | 68 (100) | 0 | 5 (100) | 0 |
| Linezolid | 68 (100) | 0 | 5 (100) | 0 |
| Teicoplanin | 68 (100) | 0 | * | * |
| Tetracycline | 38 (56) | 30 (44) | * | * |
| Gentamicin | 38 (56) | 30 (44) | 2 (40) | 3 (60) |
| Ciprofloxacin | 19 (28) | 49 (72) | * | * |
| Levofloxacin | 38 (56) | 30 (44) | * | * |
S – Sensitive; R – Resistant; IR – Intrinsically resistant; * – Not tested.
Among the gram-negative isolates (n = 53; 42%), Klebsiella pneumoniae (n = 20) was predominant, with 90% being resistant to third- and fourth-generation cephalosporins, 85% being MDR, and 70% being carbapenem resistant. Variable resistance to aminoglycosides and fluoroquinolones was observed. The second most common gram-negative species isolated was Acinetobacter baumannii (n = 14), with 100% being MDR and 80% being sensitive to colistin only (Table 4 and Figure). The remaining gram-negative isolates were sensitive to colistin, except for two isolates each of Burkholderia cepacia complex and Serratia spp., which were intrinsically resistant to colistin. However, the Burkholderia cepacia complex (n = 2), Serratia spp. (n = 2), Enterobacter cloacae complex (n = 4), and 80% of Escherichia coli and Pseudomonas aeruginosa isolates were sensitive to meropenem (Table 4).
Table (4):
Antibiotic susceptibility profile of Gram-negative isolates recovered from cases of neonatal sepsis
| Antibiotics | Antibiotic susceptibility profile of Gram-negative organisms | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacterales (N = 31) | Non-fermenters (N = 22) | |||||||||||||
| Klebsiella pneumoniae (N = 20) | Escherichia coli (N = 5) | Enterobacter cloacae complex(N = 4) | Serratia spp (N = 2) | Acinetobacter baumannii (N = 14) | Pseudomonas aeruginosa (N = 6) | Burkholderia cepacia complex (N = 2) | ||||||||
| S | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| Ampicillin | IR | IR | 0 | 5 (100) | IR | IR | IR | IR | IR | IR | IR | IR | IR | IR |
| Ceftazidime | * | * | * | * | * | * | * | * | 4 (29) | 10 (71) | 5 (84) | 1 (16) | 2 (100) | 0 |
| Ampicillin/ clavulanic acid | * | * | 2 (40) | 3 (60) | * | * | IR | IR | IR | IR | IR | IR | IR | IR |
| Piperacillin/ tazobactum | 6 (30) | 14 (70) | 4 (80) | 1 (20) | 2 (50) | 2 (50) | 0 | 2 (100) | 4 (29) | 10 (71) | 3 (50) | 3 (50) | IR | IR |
| Gentamicin | 7 (35) | 13 (65) | 2 (40) | 3 (60) | 2 (50) | 2 (50) | 1 (50) | 1 (50) | 2 (15) | 12 (85) | 5 (84) | 1 (16) | IR | IR |
| Amikacin | 2 (10) | 18 (90) | 4 (80) | 1 (20) | 3 (75) | 1 (25) | IR | IR | 3 (22) | 11 (78) | * | * | IR | IR |
| Ciprofloxacin | 2 (10) | 18 (90) | 2 (40) | 3 (60) | 2 (50) | 2 (50) | * | * | 0 | 14 (100) | 1 (17) | 5 (83) | * | * |
| Levofloxacin | 18 (90) | 2 (10) | * | * | * | * | * | * | 2 (15) | 12 (85) | 4 (67) | 2 (33) | 2 (100) | 0 |
| Ceftriaxone | 2 (10) | 18 (90) | 3 (60) | 2 (40) | 2 (50) | 2 (50) | 1 (50) | 1 (50) | 1 (8) | 13 (92) | IR | IR | IR | IR |
| Cefepime | 2 (10) | 18 (90) | 3 (60) | 2 (40) | 2 (50) | 2 (50) | 1 (50) | 1 (50) | 4 (29) | 10 (71) | 5 (84) | 1 (16) | IR | IR |
| Cefoperazone/ sulbactam | 12 (60) | 8 (40) | 4 (80) | 1 (20) | 3 (75) | 1 (25) | 1 (50) | 1 (50) | 4 (29) | 10 (71) | * | * | * | * |
| Imipenem | 6 (30) | 1470) | 2 (40) | 3 (60) | 2 (50) | 2 (50) | 2 (100) | 0 | 2 (15) | 12 (85) | 1 (16) | 5 (84) | IR | IR |
| Meropenem | 10 (50) | 10 (50) | 4 (80) | 1 (20) | 4 (100) | 0 | 2 (100) | 0 | 4 (29) | 10 (71) | 5 (84) | 1 (16) | 2 (100) | 0 |
| Ertapenem | 2 (10) | 18 (90) | 2 (40) | 3 (60) | 2 (50) | 2 (50) | 1 (50) | 1 (50) | IR | IR | IR | IR | IR | IR |
| Cotrimoxazole | 109 (50) | 10 (50) | 3 (60) | 2 (40) | 29 (50) | 2 (50) | 0 | 2 (100) | 4 (29) | 10 (71) | IR | IR | 1 (50) | 1 (50) |
| Colistin | 20 (100) | 0 | 5 (100) | 0 | 4 (100) | 0 | IR | IR | 14 (100) | 0 | 6 (100) | 0 | IR | IR |
| Minocycline | * | * | * | * | * | * | * | * | * | * | * | * | 2 (100) | 0 |
S – Sensitive; R – Resistant; IR – Intrinsically resistant;* – Not tested.
MDR organisms pose a major health risk in ICUs.6 In this study, most of the neonates in the neonatal ICU of the tertiary care hospital had a provisional diagnosis of sepsis (44%). A similar percentage (45.9%) in Egypt was reported by Shehab et al.7 In our study, 14.8% of neonates had microbiologically confirmed sepsis, with 10.5% presenting with bacteremia, an incidence rate similar to that in Southeast Europe (18%) as reported by Segal et al.8 However, these rates were lower than those reported in Nepal (40%) 9 and Southern India (23%).10 The low culture positivity in our study could be due to the prior use of antibiotics and an over-emphasis on blood culture, as the clinical presentation in this age group was subtle.
The majority of isolates recovered were gram-positive bacteria, with Staphylococcus spp. being the most common and predominant in the EOS group (p < 0.001; chi-square test, 33.49), similar to the observations made in West Africa by Fortress et al.11 However, our findings were contrary to those observed in Delhi, India by Agarwal et al.12 The predominance of Staphylococcus spp. in EOS was also observed in China by Li et al.13 By contrast, Moftian et al. observed that gram-negative isolates were the predominant causative agents of EOS in Iran.14 Staphylococcus aureus (22%) and Staphylococcus epidermidis (14%) were identified as the most common Staphylococcus spp. in our study, similar to the findings reported by Ansari et al. in Uttar Pradesh, India.15 As normal flora of the nose and skin, respectively, Staphylococcus aureus and Staphylococcus epidermidis cause bacteremia by crossing the natural defense line, especially as a result of the extensive use of invasive devices and manipulation of peripheral intravenous lines in immunologically immature neonates. Among the MDR Staphylococcus isolates identified, 72% were resistant to methicillin, with MRSA (16%) accounting for the highest proportion. By contrast, Tanu et al. observed that more than 50% of Staphylococcus aureus isolates recovered from neonates with sepsis in Delhi, India, were methicillin-resistant.16 However, in a multicenter study conducted by Kim et al. in Korea, gram-negative MDR isolates were predominant.17 MRSA, the first known widespread superbug, is associated with a high death rate and has become a serious threat, especially in ICUs, necessitating the exploration and use of new drug regimens for its eradication.18
As none of the gram-positive bacteria isolated in our study were resistant to vancomycin, linezolid, or teicoplanin, vancomycin can be used as empiric therapy for staphylococcal infections. Although the clinical effects of vancomycin and linezolid are comparable, linezolid use is associated with myelosuppression and thrombocytopenia, side effects that are leading causes of the drug being withdrawn. Similar observations were reported by Lufen et al. in a study conducted in China.19 Likewise, because limited data exist regarding the pharmacokinetics and pharmacodynamics of teicoplanin in neonates, the optimal dosing regimen remains questionable. However, researchers have proposed the routine use of therapeutic drug monitoring for optimal teicoplanin administration, as concluded by Ramos et al. in a study conducted in the United Kingdom.20
In our study, gram-negative isolates were equally responsible for both EOS and LOS. Klebsiella pneumoniae was the predominant isolate (16%) and was responsible for 7.1% of the EOS and 8.7% of the LOS cases. Similar observations were reported by Hu et al., who conducted a multicenter antimicrobial surveillance study in China.21 In our study, Acinetobacter baumannii was the most common non-fermenting gram-negative bacterium isolated (11.1%), similar to the results of Panda et al., who conducted a four-year surveillance study in Odisha, India.22 MDR Klebsiella pneumoniae was the second most resistant pathogen (13.5%) isolated in this study. By contrast, Hu et al. reported other MDR Enterobacterales as being more prevalent in their study cohort.21 The high resistance rates to third- and fourth-generation cephalosporins (90%) and carbapenems (70%) observed in the present study could be due to the circulation of extended-spectrum beta-lactamase producing strains in our region. Additionally, as 65% of the strains were resistant to gentamicin, this drug cannot be recommended as empiric therapy for treating bacteremia in our setup.
The immune systems of neonates are immature, making them more prone to hospital-acquired infections. An upsurge in bacteremia caused by MDR Klebsiella pneumoniae has been observed globally, especially in neonatal ICUs. The World Health Organization (WHO), which has included MDR Klebsiella spp. among the most dangerous superbugs, recommends treatment using only a few therapeutic options along with the detachment of invasive devices. These benefits were also suggested in a report by Ravi et al. on their study in Tamil Nadu, India.23 The success of Klebsiella pneumoniae as a pathogen can also be attributed to its hypervirulent pathotype and well-known capability to acquire antibiotic resistance determinants.21 Moreover, both of the most prevalent gram-negative organisms belonging to the ESKAPE group of pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are categorized under the “critical group” in the WHO global priority pathogen list and are almost entirely responsible for the spread of antimicrobial resistance among the strains responsible for hospital-acquired infections.18 Non-fermenting gram-negative isolates have also emerged as a major cause of nosocomial infections, being responsible for occasional outbreaks, and are the fourth most common gram-negative bacteria responsible for bacteremia. According to the Centers for Disease Control and Prevention (Atlanta, USA) nearly 80% of all reported Acinetobacter infections are caused by Acinetobacter baumannii, which is the leading agent of infections in ICUs.3 Lob et al. reported that 95% of Acinetobacter baumannii isolates collected from different centers in the USA were MDR, similar to the observations in our study (100%).24
Antimicrobial resistance is increasing to lethally high levels globally and is the most urgent public health concern worldwide.18 Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa are among the six leading agents causing high mortality rates. Their resistant strains were responsible for 3.57 million deaths in 2019, and this number may increase to 10 million by 2050 if left unchecked.25 Although none of the gram-positive and gram-negative strains isolated in this study were resistant to colistin, the observed MDR pattern of the causative agents is similar to that observed in a national multicenter study conducted in Delhi, India by investigators of the Delhi Neonatal Infection Study.12 Similar observations on neonatal sepsis in the United Kingdom were made from an international multicenter cohort study conducted by Russell et al.26 Those authors concluded that antibiotic regimens for neonatal sepsis are diverging from the WHO guidelines; hence, trials of newer empiric regimens are urgently needed to address the surge in MDR strains.26 Research on the usefulness of older antibiotics such as colistin for treating neonatal septicemia caused by MDR bacteria is warranted. Based on current data, colistin is an effective agent against MDR gram-negative bacteria, with an acceptable safety profile, indicating that it can be a useful alternative for treating MDR septicemia. However, to reduce the risk of resistance and maximize antimicrobial activity, monotherapy should be avoided, and combination therapies should be considered meticulously, as suggested by many investigators.27
Limitations
This was an observational study of a single center in northern India, which restricts the generalizability of the antibiotic resistance profiles to other locations, regardless of the substantial sample size. Moreover, clinical details and other laboratory parameters were not included, as they were not the focus of this study. Additionally, blood cultures have low sensitivity, which is further affected by prior antibiotic use in patients. Future research should focus on the application of molecular diagnostics to improve pathogen detection as well as multicenter studies to generalize the findings.
The high prevalence of MDR isolates in neonatal ICUs leads to difficulty in arriving at effective treatment decisions, thus highlighting the need for deeper investigations of the local epidemiology of causative agents of neonatal septicemia to establish empiric therapies. Given the high prevalence of gram-positive and gram-negative MDR organisms observed in our study, colistin and vancomycin can potentially be empiric therapies for neonates with suspected sepsis. Measures to reduce infection rates by undertaking regular surveillance to recognize changes in the causative agents and their drug resistance patterns should be considered. This study highlights the critical need for enhanced antimicrobial stewardship and infection control strategies in neonatal care settings. Our findings contribute to the growing body of evidence regarding the global challenge posed by MDR infectious agents and underscore the importance of continued research and policy interventions to improve neonatal outcomes.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Sher-i-Kashmir Institute of Medical Sciences, Soura, Srinagar, J&K, India, via letter No.: SIMS1131/IEC/-SKIMS/2018-244.
- Milton R, Gillespie D, Dyer C, et al. Neonatal sepsis and mortality in low-income and middle-income countries from a facility-based birth cohort: an international multisite prospective observational study. Lancet Glob Health. 2022;10(5):e661-e672.
Crossref - Celik IH, Hanna M, Canpolat FE, Pammi M. Diagnosis of neonatal sepsis: the past, present and future. Pediatr Res. 2022;91(2):337-350.
Crossref - Glossary of terms related to antibiotic resistance. https://www.cdc.gov/narms/resources/glossary.html, Accessed 01 april, 2024.
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021. https://www.who.int/ publications-detail-redirect/9789240027336, Accessed 01 april, 2024.
- Miranda S, Harahap A, Husada D, Faramarisa FN. Risk factors of multidrug-resistant organisms neonatal sepsis in Surabaya tertiary referral hospital: a single-center study. BMC Pediatr. 2024;24(1):1–8.
Crossref - Ren Z, Yang S, Han J, et al. Reduction of antibiotic use and multi-drug resistance bacteria infection in neonates after improvement of antibiotics use strategy in a level 4 neonatal intensive care unit in southern China. Eur J Clin Microbiol Infect Dis. 2023;42(1):87–98.
Crossref - Shehab El-Din EMR, El-Sokkary MMA, Bassiouny MR, Hassan R. Epidemiology of Neonatal Sepsis and Implicated Pathogens: A Study from Egypt. Biomed Res Int. 2015;2015(1):509484.
Crossref - Segal J, Hoxha M, Wien S, et al. The bacterial profile of neonatal sepsis and antibiotic use in the tertiary care NICU of Kosovo. J Pediatr Neonatal Care. 2018;8(2):105 108.
Crossref - Yadav NS, Sharma S, Chaudhary DK, et al. Bacteriological profile of neonatal sepsis and antibiotic susceptibility pattern of isolates admitted at Kanti Children’s Hospital, Kathmandu, Nepal. BMC Res Notes. 2018;11(1):301.
Crossref - Pavan Kumar DV, Mohan J, Rakesh PS, Prasad J, Joseph L. Bacteriological profile of neonatal sepsis in a secondary care hospital in rural Tamil Nadu, Southern India. J Family Med Prim Care. 2017;6(4):735-738.
Crossref - Aku FY, Akweongo P, Nyarko K, et al. Bacteriological profile and antibiotic susceptibility pattern of common isolates of neonatal sepsis, Ho Municipality, Ghana-2016. Matern Health Neonatol Perinatol. 2018;4:2.
Crossref - Agarwal, Ramesh & Sankar, Jeeva, Chaurasia, Suman. Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob Health. 2016;752-760.
Crossref - Li Z, Xiao Z, Li Z, Zhong Q, Zhang Y, Xu F. 116 cases of neonatal early-onset or late-onset sepsis: a single center retrospective analysis on pathogenic bacteria species distribution and antimicrobial susceptibility. Int J Clin Exp Med 2013;6(8):693-699.
- Moftian N, Rezaei-Hachesu P, Arab-Zozani M, et al. Prevalence of gram-negative bacteria and their antibiotic resistance in neonatal sepsis in Iran: a systematic review and meta-analysis. BMC Infect Dis. 2023;23(1):534.
Crossref - Ansari F, Banerjee T, Kumar A, Anupurba S. Coagulase-Negative Staphylococci in Neonatal Blood: How Concerning? J Lab Physicians. 2023;15(1):126-130.
Crossref - Singhal T, Rodrigues C, Soman R, et al. Treatment of MRSA infections in India: Clinical insights from a Delphi analysis. Indian J Med Microbiol. 2022;40(1):35-45.
Crossref - Kim HJ, Oh DK, Lim SY, et al. Antibiogram of multidrug-resistant bacteria based on sepsis onset location in Korea: a multicenter cohort study. J Korean Med Sci. 2023;38(10):e75.
Crossref - WHO Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 01 April, 2024.
- Duan L, Zhou Q, Feng Z, et al. A Regression Model to Predict Linezolid Induced Thrombocytopenia in Neonatal Sepsis Patients: A Ten-Year Retrospective Cohort Study. Front. Pharmacol. 2022;13:710099.
Crossref - Ramos-Martin V, Neely MN, McGowan P, et al. Population pharmacokinetics and pharmacodynamics of teicoplanin in neonates: making better use of C-reactive protein to deliver individualized therapy. J Antimicrob Chemother. 2016;71(11):3168-3178.
Crossref - Hu F, Yuan L, Yang Y, et al. A multicenter investigation of 2773 cases of bloodstream infections based on China antimicrobial surveillance network (CHINET). Front Cell Infect Microbiol. 2022;12:1075185.
Crossref - Panda SK, Nayak MK, Jena P, et al. Nonfermenting, Gram-Negative Bacilli Causing Neonatal Sepsis in Odisha, India: Four-Year Surveillance. Cureus. 2022;14(2):e22219.
Crossref - Ravi V, Vijay DD, Sujhithra A, Jayanthi S, Subramanian TK, Harish N. Neonatal Sepsis: The impact of Hypervirulent Klebsiella pneumoniain a Tertiary Care Hospital. J Pure Appl Microbiol. 2022;16(3):2035-2044.
Crossref - Lob SH, Hoban DJ, Sahm DF, et al. Regional differences and trends in antimicrobial susceptibility of acinetobacter baumannii. Int J Antimicrob Agents.2016;47:317–23.
Crossref - Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022;399(10325):629–655.
Crossref - Russell NJ, Stohr W, Plakkal N, et al. Patterns of antibiotic use, pathogens, and prediction of mortality in hospitalized neonates and young infants with sepsis: A global neonatal sepsis observational cohort study (NeoOBS). PLOS Med. 2023;20(6):e1004179.
Crossref - Baltogianni M, Dermitzaki N, Kosmeri C, Serbis A, Balomenou F, Giapros V. Reintroduction of Legacy Antibiotics in Neonatal Sepsis: The Special Role of Fosfomycin and Colistin. Antibiotics2024;13(4):333.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.