ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is an expedient Gram-negative bacterium, which is characterized by its ability to acquire antimicrobial resistance. In this study, 56 unrepeatable carbapenem-resistant P. aeruginosa isolates were gathered from various clinical sources from hospitals in Cairo and Mansoura universities. The isolates exhibited diminished susceptibility towards carbapenems, quinolones, aminoglycosides and chloramphenicol by using disc diffusion method. Carbapenemase production was confirmed among the isolates, where all the 56 P. aeruginosa isolates harboured carbapenemase genes including blaVIM (43 isolates), blaKPC (38 isolates), blaNDM-1 (17 isolates), blaIMP (16 isolates) and blaOXA-48 (15 isolates). Among the isolates, 13 carried only one carbapenemase gene, while 43 isolates carried multiple carbapenemase genes. MCR-1 production was confirmed in 10 of the tested isolates by detecting the mcr-1 gene encoding for the colistin resistance. Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) evaluation showed that the tested isolates were unrelated to each other. Therefore, this study rises the danger of emergence of MDR P. aeruginosa resistant to carbapenems coupled with other antimicrobials including colistin, which is regarded as the last reservoir for the management of infections caused by MDR Gram-negative pathogens. Early inspection of resistance patterns in MDR organisms is an important tool to control and prevent infections via limiting the spread of these pathogens.
Carbapenemase, Colistin, Egypt, Multidrug-resistance, Pseudomonas aeruginosa
Pseudomonas aeruginosa is a threatening human pathogen that is usually linked to nosocomial infections.1,2 Commonly, this pathogen is multidrug-resistant (MDR) due to its extraordinary capacity to acquire resistance to a wide range of antimicrobials.3 Usually, carbapenems are considered the first treatment choice against infections brought about by P. aeruginosa. Because of the widespread use of carbapenems, there has been a global increase in reports regarding P. aeruginosa carbapenem resistant isolates.4 It is also notable that isolates of P. aeruginosa have started to depict resistance to the antibiotics that are used as a last choice for therapy including colistin.5
Indeed, the primary mechanism for pathogens to resist carbapenems is the production of carbapenemases, which are mainly plasmid mediated and considered as a type of β-lactamases.6 Carbapenemases have a wide range of hydrolytic capabilities against antimicrobials including carbapenems, cephalosporins and penicillins.7,8 Three major types of carbapenemases have been identified to be the source of nosocomial epidemics in P. aeruginosa involving the KPC type (class A serine enzymes), the IMP, VIM and NDM-1 types (class B metallo-β-lactamases (MBLs) or metal enzymes) and the OXA-48 type (class D enzymes).7-11 Class B enzymes are MBLs with zinc in the active site, while class A and D enzymes have a serine-based hydrolytic mechanism.8-10,12
The most significant developing mechanisms of β-lactam resistance in P. aeruginosa are those arbitrated by extended-spectrum β-lactamases (ESBLs) and MBLs. β-lactamases inhibitors do not inhibit MBLs, which have a significant capacity to hydrolyze carbapenems efficiently and other β-lactams.10,11,13,14 The KPC type is primarily present on pKpQIL plasmid, which occurs in Klebsiella pneumonia.15 The OXA-48 type is frequently found on carbonen 62-kb IncL/M conjugative plasmids of Acinetobacter baumannii.12
Notably, polymyxins are typically considered the final treatment line against MDR Gram-negative bacteria. The polycationic peptide called colistin (also named to as polymyxin E) binds to anionic lipopolysaccharide molecules in the outer membrane of Gram-negative cell walls and disrupts them by competing with Ca2+ and Mg2+ cations for these molecules.16,17 Unfortunately, colistin resistance has recently developed because of the drug misuse.16 The horizontal transfer of phosphoethanolamine transferase enzyme-encoding gene mcr-1 was discovered in 2015 and it has a key role in the colistin resistance.18 This enzyme has the ability to modify the lipopolysaccharides of the outer membrane lipid A of Gram-negative pathogens.17,19
The horizontal transfer of plasmid-mediated resistance has two risks. First, the plasmids can confer the resistance to a variety of drugs. Second, plasmids have a stronger capacity to propagate resistance among bacteria than natural mutation.4-6 The rise of carbapenem-resistant pathogens all over the world with identical mobile genetic elements suggests that genes for carbapenemases have been spread horizontally.6 The MBL-producing genes blaVIM, blaNDM-1 and blaIMP as well as blaKPC (class A) and blaOXA-48 (class D) genes can transfer horizontally via plasmids and can spread quickly to other bacteria.4,5,7,8,10,11,20 The same case is for mcr-1, which is plasmid mediated and has the ability to spread quickly over the globe.17,19
Based on this context, the objective of the present investigation was to estimate the dominance of MBLs (VIM, NDM-1 and IMP) as well as class serine enzymes (the KPC type and OXA-48 type), which are responsible for carbapenemases activity in the isolates of P. aeruginosa. Moreover, to investigate the prevalence of MCR-1 among the isolates. After all, the clonal clustering of the tested isolates was established utilizing (ERIC)-PCR.
Bacterial isolates and media
This study was carried out after the approval of the research ethics committee of Faculty of Pharmacy, Tanta University, Egypt (code: TP / RE /12-21-M-002). The current study included 56 unrepeatable carbapenem-resistant P. aeruginosa isolates. These isolates were gathered from several clinical sources from hospitals in Cairo and Mansoura universities. The examined isolates were collected, identified and stored using the standard microbiological procedures.21 All isolates were cultured at 37°C in Luria Bertani medium (LB broth; tryptone 1% w/v, yeast extract 0.5% w/v and NaCl 1.0% w/v), otherwise specified, and stored in 50% glycerol/LB broth at -80°C. During the culture and sensitivity testing, the isolates were cultured on modified Muller-Hinton agar (MHA) supplemented with all antimicrobials, except for colistin as described before.22 For colistin, to improve its diffusion, the modified MHA containing 30% agar was used.
Antimicrobial sensitivity testing
Antimicrobial susceptibility tests were performed on the isolates against several antimicrobial classes (Oxoid, UK) including β-lactams (sulbactam, cefepime, cefoperazone, ceftazidime, carbenicillin, ceftriaxone, cefotaxime, ampicillin, meropenem and imipenem), quinolones (levofloxacin, ciprofloxacin, norfloxacin and nalidixic Acid), aminoglycosides (amikacin, gentamicin and tobramycin), chloramphenicol and colistin by using disc diffusion method.23 Antimicrobial susceptibility was established using the clinical and laboratory standards institute (CLSI) 2019 and European committee on antimicrobial susceptibility testing (EUCAST) recommendations.6,22,24 As a control, Escherichia coli ATCC 25922 standard strain was used during these experiments.
Carbapenemase phenotypic examination
Modified Hodge test (MHT) was carried out to test carbapenemase production as described before.25 A commercial disc containing 10 μg of imipenem was placed in the middle of a MHA plate that had been inoculated with E. coli ATCC 25922. The tested P. aeruginosa isolates were considered β-lactamase positive if they enabled E. coli ATCC 25922 strain to resist the imipenem giving a cloverleaf-like indentation. During these experiments, K. pneumoniae ATCC BAA-1705 and ATCC BAA-1706 strains were used as positive and negative controls, respectively.
EDTA synergistic test was used to evaluate the MBLs activity as indicated before.26 This test involves challenging the tested isolate with one disc containing anhydrous EDTA (292 μg) (Sigma Chemicals in St. Louis, MO) and two imipenem discs (10 μg each). The discs are spaced 25 mm apart in the MHA plate. Positive MBL production was demonstrated by an increase in the inhibition zone width of more than 4 mm around the imipenem-EDTA disc in comparison to the imipenem disc alone. As a control, E. coli ATCC 25922 standard strain was used during these experiments.
Furthermore, KPC enzyme synthesis evaluation was established by the boronic acid test as specified before.27 Antimicrobial discs (cefepime, meropenem or imipenem) and a KPC inhibitor (400 μg benzene boronic acid; Sigma-Aldrich, Steinheim, Germany) were employed in this assay. The tests were carried out by inoculating a MHA plate with the tested isolate in the presence of antimicrobial discs with or without boronic acid. Antimicrobial-boronic acid disk is compared to the antibiotic disc alone, and KPC enzyme production is considered positive if the diameter of the inhibitory zone around the former increases by 5 mm or more. The standard strain E. coli ATCC 25922 was used in this test to establish quality control.
Polymerase chain reaction (PCR)
The QIA amp® DNA miniprep kit (Qiagen, Germany) was utilized to extract plasmid DNA from P. aeruginosa isolates. Screening for the existence of β-lactamases genes involving blaVIM, blaIMP, blaNDM-1, blaKPC and blaOXA-48 and colistin resistance gene (mcr-1) was established by PCR using the primer sets listed in Table 1.28-32 The PCR reactions were established in the volume of 25 μL. Each PCR reaction consisted of 3 μL of template DNA, 1 μL forward primer (10 μM), 1 μL of reverse primer (10 μM), 12.5 μL Dream Taq PCR master mix 2x (Fermentas, USA) and 7.5 μL nuclease free water. Tubes with no template DNA were used as a negative control. The PCR cycling was established as follows; primary denaturation for 5 min at 95°C, followed by 35 cycles of (denaturation for 30 s at 95°C, annealing for 30 s at 52°C for blaVIM, blaIMP, blaNDM-1 and mcr-1, while at 48°C and 58°C for blaKPC and blaOXA-48, respectively and extension for 1 min at 72°C) and last extension for 5 min at 72°C.
Table (1):
Oligonucleotide primers used in this study.
Gene name |
Primer Type |
Sequence |
Amplicon size (bp) |
Reference |
|---|---|---|---|---|
blaVIM |
F |
5`… ATTGGTCTATTTGACCGCGTC |
780 |
29, 31 and 32 |
R |
5`… TGCTACTCAACGACTGAGCG |
|||
blaIMP |
F |
5`… CATGGTTTGGTGGTTCTTGT |
488 |
29, 31 and 32 |
R |
5`… ATAATTTGGCGGACTTTGGC |
|||
blaKPC |
F |
5`… CGTTGACGCCCAATCC |
390 |
29, 31 and 32 |
R |
5`… ACCGCTGGCAGCTGG |
|||
blaOXA-48 |
F |
5`…ATGCGTGTATTAGCCTTATCGGC |
770 |
29, 31 and 32 |
R |
5`…ACTTCTTTTGTGATGGCTTGGCGCA |
|||
blaNDM-1 |
F |
5`… GGTTTGGCGATCTGGTTTTC |
621 |
30 |
R |
5`… CGGAATGGCTCATCACGATC |
|||
mcr-1 |
F |
5`… CGGTCAGTCCGTTTGTTC |
309 |
28 |
R |
5`… CTTGGTCGGTCTGTAGGG |
|||
ERIC-2 |
5`… AAGTAAGTGACTGGGGTGAGCG |
33 |
F: Forward, R: Reverse
Results of PCR were inspected using gel electrophoresis, where the products were run on 1.2% agarose gel and visualized utilizing ethidium bromide (MP biomedicals, France). The size of the obtained amplicons was matched to DNA ladder (Thermo Scientific, UK). The tested gene was considered positive if a single sharp band of its expected size appeared beside the matched band of the DNA ladder reflecting its size.
Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR)
The clonal relatedness of the P. aeruginosa isolates was done using ERIC-PCR. The banding profile was generated using the oligonucleotide primer ERIC-2 (Table 1)33 according to the method previously described.34 The resultant patterns attained from ERIC-PCR were construed using the software Past® (version 4.01).35 The similarities among the fingerprints were calculated based on Pearson correlation (optimization, 1%; position tolerance, 1%). By using the UPGMA algorithm, the fingerprints were sorted based on their similarity to generate their corresponding dendrograms.36 Isolates with a resemblance of more than 85% were deemed clonal.
Clinical features and antimicrobial sensitivity patterns of the tested isolates
All tested P. aeruginosa were carbapenemase-producing isolates and harboured carbapenemase genes. According to the clinical source as shown in Table 2, 22 isolates (39.28%) were gathered from urine, 13 isolates (23.21%) from wounds, 9 isolates (16.1%) from burns, 6 isolates (10.71%) from sputum, 4 isolates (7.14%) from ear swabs and 2 isolates (3.57%) from eye infections. In a 1:1 ratio, samples were drawn from both males and females. Regarding the antimicrobial susceptibility patterns of the tested isolates as shown in Table 3, the highest resistance percentage was observed with sulbactam/amoxicillin, cefoperazone, cefotaxime, ampicillin and nalidixic acid, where 100% of the isolates were resistant. Moreover, the resistance percentage to ceftazidime and ceftriaxone was 94.6%, followed by carbenicillin (91.1%) and both tobramycin and gentamicin (80.35%). The resistance percentages to meropenem and imipenem were 96.4% and 75%, respectively. On the other hand, colistin was shown to have the lowest resistance rate (17.8%), while amikacin and norfloxacin resistance was (44.6%). In addition, the resistance percentages to levofloxacin, ciprofloxacin and chloramphenicol were 55.3%, 46.4% and 62.5%, respectively. The examined isolates U25 and B56 showed complete resistance to all antimicrobials used in this study.
Table (2):
Distribution of carbapenem and colistin resistance genes in P. aeruginosa tested isolates and their phenotypic detection.
Isolate code |
Source |
Sex |
Ward |
Infection |
blaVIM |
blaKPC |
blaNDM-1 |
blaIMP |
blaOXA-48 |
mcr-1 |
MHT |
EDTA synergistic test |
Boronic acid test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
U5 |
Urine |
F |
UNC |
UTI |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
U6 |
Urine |
F |
MUH |
UTI |
+ |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
U7 |
Urine |
F |
CUH |
UTI |
+ |
+ |
+ |
+ |
– |
– |
+ |
+ |
+ |
U8 |
Urine |
F |
MUH |
UTI |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
S9 |
Sputum |
F |
MUH |
RTI |
+ |
+ |
+ |
+ |
– |
– |
+ |
+ |
+ |
U10 |
Urine |
F |
UNC |
UTI |
+ |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
U11 |
Urine |
M |
MUH |
UTI |
+ |
+ |
+ |
+ |
– |
– |
+ |
+ |
+ |
U12 |
Urine |
M |
UNC |
UTI |
+ |
+ |
– |
+ |
– |
– |
+ |
+ |
+ |
U13 |
Urine |
M |
ICU |
UTI |
+ |
– |
– |
– |
– |
– |
– |
+ |
– |
U14 |
Urine |
M |
MUH |
UTI |
+ |
– |
+ |
– |
– |
– |
– |
+ |
– |
U23 |
Urine |
F |
MUH |
UTI |
+ |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
U24 |
Urine |
F |
UNC |
UTI |
+ |
+ |
– |
+ |
– |
– |
+ |
+ |
+ |
U25 |
Urine |
M |
MUH |
UTI |
+ |
+ |
– |
+ |
– |
+ |
– |
+ |
+ |
U26 |
Urine |
F |
CUH |
UTI |
+ |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
U35 |
Urine |
F |
MUH |
UTI |
+ |
– |
+ |
– |
– |
+ |
– |
+ |
– |
U36 |
Urine |
F |
UNC |
UTI |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
U40 |
Urine |
F |
CUH |
UTI |
+ |
+ |
– |
– |
+ |
– |
+ |
+ |
+ |
W41 |
wounds |
F |
ICU |
WI |
+ |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
W42 |
wounds |
F |
ICU |
WI |
+ |
– |
– |
+ |
– |
– |
+ |
+ |
– |
W43 |
wounds |
F |
ICU |
WI |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
W44 |
wounds |
F |
ICU |
WI |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
W45 |
wounds |
F |
CUH |
WI |
– |
+ |
– |
– |
– |
+ |
– |
– |
+ |
W46 |
wounds |
M |
ICU |
WI |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
W50 |
wounds |
M |
BCC |
WI |
– |
+ |
– |
– |
+ |
– |
– |
– |
+ |
W51 |
wounds |
M |
BCC |
WI |
– |
+ |
– |
+ |
– |
– |
+ |
+ |
+ |
W52 |
wounds |
M |
BCC |
WI |
– |
– |
– |
+ |
– |
– |
– |
+ |
– |
W54 |
wounds |
M |
MUH |
WI |
– |
– |
– |
+ |
– |
– |
+ |
+ |
– |
W55 |
wounds |
M |
MUH |
WI |
+ |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
B56 |
Burns |
M |
BCC |
BI |
+ |
+ |
+ |
– |
+ |
+ |
– |
+ |
+ |
B62 |
Burns |
M |
BCC |
BI |
– |
– |
– |
– |
+ |
– |
+ |
– |
– |
B63 |
Burns |
M |
BCC |
BI |
– |
– |
+ |
– |
+ |
– |
+ |
+ |
– |
B65 |
Burns |
M |
BCC |
BI |
– |
+ |
+ |
– |
+ |
+ |
– |
+ |
+ |
S66 |
Sputum |
M |
CH |
RTI |
– |
– |
– |
– |
+ |
– |
+ |
– |
– |
S68 |
Sputum |
M |
CH |
RTI |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
S69 |
Sputum |
F |
MIH |
RTI |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
S71 |
Sputum |
M |
MIH |
RTI |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
B73 |
Burns |
F |
BCC |
BI |
– |
+ |
+ |
+ |
– |
– |
– |
+ |
+ |
W74 |
wounds |
F |
BCC |
WI |
+ |
+ |
– |
– |
+ |
– |
+ |
+ |
+ |
B75 |
Burns |
M |
BCC |
BI |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
U76 |
Urine |
F |
UNC |
UTI |
+ |
+ |
– |
+ |
+ |
– |
+ |
+ |
+ |
U78 |
Urine |
F |
CUH |
UTI |
+ |
+ |
– |
– |
+ |
– |
– |
+ |
+ |
U80 |
Urine |
F |
MUH |
UTI |
+ |
+ |
– |
– |
+ |
+ |
+ |
+ |
+ |
EY81 |
Eye |
M |
OC |
EI |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
S82 |
Sputum |
M |
MUH |
RTI |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
EY83 |
Eye |
M |
OC |
EI |
+ |
+ |
– |
– |
– |
– |
+ |
+ |
+ |
B84 |
Burns |
M |
BCC |
BI |
+ |
+ |
+ |
– |
+ |
+ |
– |
+ |
+ |
U85 |
Urine |
M |
MUH |
UTI |
+ |
+ |
+ |
– |
+ |
– |
– |
+ |
+ |
U86 |
Urine |
F |
MUH |
UTI |
+ |
+ |
– |
– |
– |
+ |
+ |
+ |
+ |
E87 |
Ear |
F |
MUH |
OM |
+ |
+ |
+ |
– |
+ |
– |
– |
+ |
+ |
E90 |
Ear |
F |
CUH |
OM |
+ |
+ |
+ |
– |
+ |
– |
– |
+ |
+ |
B95 |
Burns |
M |
BCC |
BI |
– |
+ |
– |
– |
– |
– |
– |
– |
+ |
W96 |
wounds |
F |
MUH |
WI |
– |
– |
+ |
– |
– |
– |
– |
+ |
– |
E97 |
Ear |
F |
MUH |
OM |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
U98 |
Urine |
M |
MUH |
UTI |
+ |
– |
– |
+ |
– |
– |
+ |
+ |
– |
E99 |
Ear |
M |
CUH |
OM |
– |
+ |
– |
– |
– |
– |
+ |
– |
+ |
B100 |
Burns |
M |
BCC |
BI |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
B: Burns, BCC: Burns and cosmetics center, BI: Burn infection, CH: Chest hospital, CUH: Cairo university hospital, E: Ear swabs, EI: Eye infection, EY: Eye, F: Female, ICU: Infection control unit, M: Male, MHT: Modified Hodge test, MIH: Mansoura international hospital, MUH: Mansoura university hospital, OC: Ocular center, OM: Otitis media, RTI: Respiratory tract infection, S: Sputum, U: Urine, UNC: Urology and nephrology center, UTI: Urinary tract infection, W: Wound, WI: Wound infection
Phenotypic and genotypic detection of carbapenemases and MCR-1 among the tested isolates
According to Table 2, 31 isolates (55.35%) were MHT positive. Furthermore, the EDTA synergistic test was positive in 50 isolates (89.23%). In addition, the boronic acid test gave positive findings with 38 isolates (67.85%).
Table (3):
Antimicrobial susceptibility patterns of P. aeruginosa tested isolates.
| Isolate code | Antimicrobial class | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-lactams | Quinolones | Amino-glycosides | Lipo-peptide | ||||||||||||||||
| SAM | FEP | CFP | CAZ | CAR | CRO | CTX | AMP | MEM | IPM | LEV | CIP | NOR | NA | AK | TOB | CN | CT | C | |
| U5 | R | S | R | S | S | R | R | R | S | R | S | S | S | R | S | S | S | S | S |
| U6 | R | R | R | I | S | R | R | R | R | S | S | S | S | R | S | S | S | S | R |
| U7 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | R | R | S | R |
| U8 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| S9 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| U10 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | S | R |
| U11 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| U12 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| U13 | R | R | R | R | R | R | R | R | R | R | R | I | I | R | S | R | R | S | R |
| U14 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | R | R | S | R |
| U23 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| U24 | R | R | R | R | R | R | R | R | R | I | R | R | R | R | R | R | R | S | R |
| U25 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| U26 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| U35 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | I | R | R | R | R |
| U36 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | I | R | S | R |
| U40 | R | R | R | R | R | R | R | R | R | R | S | R | R | R | I | R | R | S | R |
| W41 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | R | R | S | R |
| W42 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | R | R | S | R |
| W43 | R | R | R | R | R | R | R | R | R | I | R | R | R | R | S | R | R | S | R |
| W44 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | S | R | S | R |
| W45 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S |
| W46 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | I | R | S | R |
| W50 | R | R | R | R | R | R | R | R | S | R | S | S | S | R | R | R | R | S | R |
| W51 | R | I | R | R | R | R | R | R | R | S | R | R | R | R | S | S | S | S | R |
| W52 | R | S | R | R | S | I | R | R | R | S | S | S | S | R | S | S | S | S | S |
| W54 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R |
| W55 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | I | R | R | S | R |
| B56 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| B62 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | S | R |
| B63 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | S | R | R | S | R |
| B65 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | S | R | R | R | R |
| S66 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | R | R | S | R |
| S68 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | I | R | S | R |
| S69 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | I | R | S | S | R |
| S71 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | R | S | R | S | R |
| B73 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | S | R | R | S | I |
| W74 | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | R | R | S | I |
| B75 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | S | R | R | R | I |
| U76 | R | R | R | R | R | R | R | R | R | R | R | S | S | R | S | R | R | S | I |
| U78 | R | I | R | R | R | R | R | R | R | R | R | I | S | R | I | R | R | S | I |
| U80 | R | R | R | R | R | R | R | R | R | R | I | I | S | R | S | R | R | R | I |
| EY81 | R | I | R | R | R | R | R | R | R | R | I | S | S | R | S | R | R | R | I |
| S82 | R | R | R | R | R | R | R | R | R | R | I | I | S | R | S | R | S | S | I |
| EY83 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | S | R | S | S | I |
| B84 | R | R | R | R | R | R | R | R | R | R | R | I | S | R | S | R | S | R | I |
| U85 | R | R | R | R | R | R | R | R | R | R | R | I | S | R | R | R | S | S | I |
| U86 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | R | R | S | R | I |
| E87 | R | S | R | R | R | I | R | R | R | I | R | R | R | R | S | R | R | S | R |
| E90 | R | S | R | I | S | R | R | R | R | I | S | S | S | R | S | S | S | S | R |
| B95 | R | R | R | R | R | R | R | R | R | R | S | I | S | R | S | R | R | S | I |
| W96 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | S | R | R | S | I |
| E97 | R | R | R | R | I | I | R | R | R | S | R | R | S | R | R | R | R | S | I |
| U98 | R | R | R | R | R | R | R | R | R | R | I | I | S | R | S | R | R | S | I |
| E99 | R | R | R | R | R | R | R | R | R | R | S | S | S | R | S | R | R | S | I |
| B100 | R | R | R | R | R | R | R | R | R | R | I | S | S | R | S | R | R | S | I |
| Total | 100% | 87.50% | 100% | 94.60% | 91.10% | 94.60% | 100% | 100% | 96.40% | 75% | 55.30% | 46.40% | 44.60% | 100% | 44.60% | 80.35% | 80.35% | 17.80% | 62.50% |
AK: Amikacin, AMP: Ampicillin, B: Burns, C: Chloramphenicol, CAR: Carbenicillin, CAZ: Ceftazidime, CFP: Cefoperazone, CIP: Ciprofloxacin, CN: Gentamicin, CRO: Ceftriaxone, CT: Colistin, CTX: Cefotaxime, E: Ear swabs, EY: Eye, FEP: Cefepime, IPM: Imipenem, LEV: Levofloxacin, MEM: Meropenem, NA: Nalidixic Acid, NOR: Norfloxacin, S: Sputum, SAM: Sulbactam, TOB: Tobramycin, U: Urine, W: Wound
PCR detection of genes encoding carbapenemase and MCR-1 confirmed the phenotypic outcomes as shown in Table 2, blaVIM, blaKPC, blaNDM-1, blaIMP and blaOXA-48 genes were detected in 43 (76.7%), 38 (67.7%), 17 (30.3%), 16 (28.5%) and 15 (26.7%) isolates, respectively. blaVIM was the most frequent gene in the tested isolates. On the other hand, blaOXA-48 was the least common gene. In addition, 43 isolates (76.78%) harboured more than one carbapenemase gene as shown in Table 4, while 13 (23.2%) isolates harboured only one gene as shown in Table 5. Regarding mcr-1 gene, it was detected in 10 P. aeruginosa carbapenemase-producing isolates, which were collected from urine (4 specimens), wound (1 specimen), burns (4 specimens) and eye infection (1 specimen) (Tables 2, 4 and 5).
Table (4):
P. aeruginosa isolates that harbour more than one carbapenem-resistant gene accompanied with the colistin resistant gene, mcr-1.
Carbapenem resistance genes and mcr-1 |
Isolate codes |
Total isolates (56 isolates) |
|---|---|---|
blaVIM + blaKPC+ blaNDM-1+ blaIMP |
U7+S9+U11 |
3 (5.35%) |
blaVIM + blaKPC + blaNDM-1+ blaOXA-48 |
E87+E90+U85 |
3 (5.35%) |
blaVIM + blaKPC + blaNDM-1+ blaOXA-48 + mcr-1 |
B56+B84 |
2 (3.57%) |
blaVIM + blaKPC + blaIMP + blaOXA-48 |
U76 |
1 (1.78%) |
blaVIM + blaKPC + blaIMP |
U6+U10+U12+U23+U24 |
5 (8.92%) |
blaVIM + blaKPC + blaIMP + mcr-1 |
U25 |
1 (1.78%) |
blaVIM + blaKPC + blaOXA-48 |
W74+U78+U40 |
3 (5.35%) |
blaVIM + blaKPC + blaOXA-48 + mcr-1 |
U80 |
1 (1.78%) |
blaKPC + blaNDM-1+ blaOXA-48 + mcr-1 |
B65 |
1 (1.78%) |
blaKPC + blaNDM-1+ blaIMP |
B73 |
1 (1.78%) |
blaVIM + blaKPC |
U5+U8+U26+U36+W41+ |
9 (16.1%) |
W55+S82+EY83+B100 |
||
blaVIM + blaKPC + mcr-1 |
B75+EY81+U86 |
3 (5.35%) |
blaVIM + blaNDM-1 |
U14+W44+S68+S69 |
4 (7.1%) |
blaVIM + blaNDM-1 + mcr-1 |
U35 |
1 (1.78%) |
blaVIM + blaIMP |
W42+U98 |
2 (3.57%) |
blaKPC + blaIMP |
W51 |
1 (1.78%) |
blaKPC + blaOXA-48 |
W50 |
1 (1.78%) |
blaNDM-1 + blaOXA-48 |
B63 |
1 (1.78%) |
Total |
43 (76.7%) |
Table (5):
P. aeruginosa isolates that harbour only one carbapenem-resistant gene accompanied with the colistin resistant gene, mcr-1.
Carbapenem resistance gene and mcr-1 |
Isolate codes |
Total isolates (56 isolates) |
|---|---|---|
blaVIM |
W46+E97+U13+ |
5 (8.92%) |
W43+S71 |
||
blaKPC |
B95+ E99 |
2 (3.57%) |
blaKPC + mcr-1 |
W45 |
1 (1.78%) |
blaNDM-1 |
W96 |
1 (1.78%) |
blaIMP |
W52+W54 |
2 (3.57%) |
blaOXA-48 |
B62+S66 |
2 (3.57%) |
Total |
13 (23.2%) |
Genetic relatedness of the P. aeruginosa tested isolates
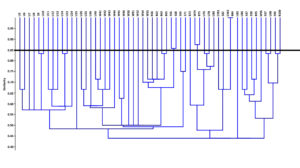
ERIC-PCR analysis of the isolates, as shown in Figure, revealed that they are unrelated to each other, except for the isolates (EY83 and B84), (S66 and S68) and (W74 and B75), which have more than 85% similarity and were considered clonal.
Figure. Dendrogram of ERIC-PCR exhibiting the clonal relatedness of P. aeruginosa isolates. The resulting patterns obtained from ERIC-PCR were interpreted using the software Past® (version 4.01). The similarities between the fingerprints were calculated based on Pearson correlation (optimization, 1%; position tolerance, 1%), and the fingerprints were grouped according to their similarities by using UPGMA algorithm to generate their corresponding dendrograms. Isolates with > 85% similarity were considered clonal. ERIC-PCR, enterobacterial repetitive intergenic consensus polymerase chain reaction
P. aeruginosa is one of the most prevalent Gram-negative opportunistic bacteria that can result in nosocomial infections. The scenario becomes worse if the infections are linked to MDR pathogens, which limits the treatment options 37. Indeed, the resistance of Gram-negative pathogens, including P. aeruginosa, to carbapenems is a global health concern.10
This study found that each isolate of the collected P. aeruginosa was at least resistant to one carbapenem (imipenem or meropenem). Moreover, 50 isolates (89.23%) were MBL producers. Previous reports showed similar results, where a study indicated that 12 P. aeruginosa isolates of 80 (26.25%) were carbapenemase producers.10 According to another study, 14 P. aeruginosa isolates of 114 (12.2%) were carbapenem resistant. Thirteen isolates of these 14 isolates (11.4%) exhibited the MBL phenotype.11 Another two studies established in Brazil and Korea demonstrated that 43.9% and 92.7% of P. aeruginosa isolates were resistant to carbapenem, respectively.20,38 In addition, a study that was carried out in Canada depicted that 228 patients were infected with imipenem resistant P. aeruginosa. This study demonstrated that 98 isolates were MBL producers.39 In addition, a study conducted in Iran indicated that 110 isolates out of 122 (90 %) were imipenem resistant and MBL producers.40
Indeed, MHT is a confirmatory phenotypic test for the inspection of P. aeruginosa carbapenemase synthesis. However, it should be noted that the sensitivity and specificity of MHT for detecting carbapenemase synthesis are debatable. Although all the tested isolates generated carbapenemases, 31 isolates (55.3%) were MHT positive and the remaining 25 isolates (44.6%) showed negative results. Several investigations have also indicated that the MHT may produce false positive or negative results during the detection of carbapenemase synthesis in Gram-negative pathogens.41-43 Unlike MHT, EDTA synergistic test used to inspect MBL synthesis, including VIM, NDM-1 and IMP, was shown to be significantly receptive, because all MBL P. aeruginosa producing isolates (50 isolates) were EDTA synergistic test positive. Furthermore, the boronic acid test was extremely responsive for detecting KPC enzyme synthesis, because of giving positive results with all KPC-positive P. aeruginosa isolates (38 isolates). Therefore, our findings suggest that the EDTA synergistic test and the boronic acid test can be routinely used in microbiology laboratories to check MBL and KPC producing P. aeruginosa isolates.
Former reports stated that the blaVIM gene is the most common produced gene in carbapenem-resistant P. aeruginosa.10,11,13,14 This is in accordance with this study, where blaVIM was harboured by 46 P. aeruginosa isolates (76.78%) as the most common MBL. In a study conducted in Egypt, blaVIM gene was found in 8 (57%) isolates out of 14 P. aeruginosa imipenem resistant isolates 11. Moreover, this gene was found in 19 (55.88%) isolates out of 34 carbapenem-resistant P. aeruginosa isolates in another report.10 In two Iranian studies,40 and44 the prevalence rates of the blaVIM gene were reported to be 1.6% and 55%, respectively. Another study established in Canada indicated that 90 (39.47%) isolates out of 228 imipenem resistant P. aeruginosa isolates harboured blaVIM gene. Moreover, during the year 2003, a nosocomial outbreak was caused by a cluster of blaVIM producing strains.39 Another study in Poland depicted a higher incidence of P. aeruginosa strains harbouring the blaVIM gene (68%).45
In this study, blaIMP gene was found in 16 P. aeruginosa isolates (28.57%). In Egypt, a study demonstrated that 5 isolates out of 14 imipenem resistant P. aeruginosa isolates (35%) harboured blaIMP gene.11 Other studies found lower incidence rates of blaIMP including 1.75% in Canada,39 and 3% in Iran.44 Another study in Iran, on the other hand, depicted a greater prevalence of blaIMP (55%) through the tested isolates of P. aeruginosa.40
In Egypt, the first report of P. aeruginosa harbouring blaNDM-1 was published in 2014,14 where the authors showed that 2 P. aeruginosa isolates out of 33 (6%) were carbapenem-resistant and harboured blaNDM-1. Moreover, another report showed a greater incidence of P. aeruginosa isolates that are resistant to carbapenem carrying blaNDM-1.13 According to this study, blaNDM-1 gene was found in 17 P. aeruginosa carbapenems and carrying isolates (30.4%) and this percentage is high in relation to the previous reports.
Regarding blaKPC, in Egypt, a study demonstrated that this gene was found in 1 isolate (2.9%) out of 34 P. aeruginosa carbapenemase-producing isolates, while blaOXA-48 gene was not found in all 34 carbapenemase-producing isolates.10 In the current study, blaKPC and blaOXA-48 occurred in 38 (67.85%) and 15 (26.78%) isolates, respectively. These incidence percentages of both genes are much higher than the previous reports. Another study conducted in Iran indicated that blaKPC presented in 13% of 108 P. aeruginosa isolates,41 while in Puerto Rico, 99 (4.1%) out of 2415 P. aeruginosa isolates harboured blaKPC gene 46. Moreover, a study in Sudan reported that 60% of P. aeruginosa isolates carried blaOXA-48 gene.47
According to this study, 43 (76.78%) P. aeruginosa isolates carried more than one carbapenem resistance gene. Compared to another report in Egypt, this percentage is very high. Previous report showed that 1 isolate out of 34 P. aeruginosa carbapenemase-producing isolates harboured both blaVIM and blaKPC.10 Another study depicted that only 4 (28.5%) isolates out of 14 P. aeruginosa carbapenemase-producing isolates carried both blaVIM and blaIMP.11 This gives an indication for the prevalence of carbapenem resistance genes that increased and became a serious problem in Egypt.
According to the data of this study, the lowest percentage of resistance among the tested P. aeruginosa isolates was recorded for colistin (10%). Given the fact that colistin has the potential to be nephrotoxic, it should only be used as a last choice for serious infections that cannot be cured with antimicrobial combinations.48 In this study, all P. aeruginosa colistin resistant isolates harboured mcr-1 gene. These data revealed high colistin resistance percentage compared to previous studies that showed complete sensitivity to colistin 10 and another study that demonstrated that only 1 isolate from 66 P. aeruginosa isolates was resistant to colistin.28
In the present study, the homology analysis (using ERIC-PCR) revealed that plasmid-mediated carbapenemases and colistin resistance genes could be easily transmitted across the P. aeruginosa isolates. Most of the tested isolates were unrelated to each other; however, they all included one or more carbapenemase gene. According to these findings, it can be concluded that P. aeruginosa isolates can easily share plasmids that encode antimicrobial resistance markers. These findings support earlier studies that described the horizontal spread of plasmids encoding carbapenemases as well as MCR-1 among Gram-negative bacteria.4,10,11,13,14,20,38,41,47,49
Overall, one of the biggest threats to the healthcare systems globally is the appearance of MDR pathogens. This study demonstrated the presence of MDR P. aeruginosa isolates that are resistant to both carbapenems and colistin as well as other antimicrobials. Early inspection of MDR P. aeruginosa isolates with any diminished susceptibility to the carbapenems as well as colistin is essential for the choice of the most proper antimicrobial treatment and the application of effective infection management protocols. Increasing the health awareness and the appropriate use of antimicrobials, particularly carbapenems and cephalosporins, as well as the minimalistic use of colistin may help to prevent the emergence of such resistance patterns.
ACKNOWLEDGMENTS
The authors would like to thank Microbiology and Immunology Departments of Faculty of Pharmacy in Horus University, Tanta University and Mansoura University, Egypt, for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
FS, TEB and AE designed the study. KMA collected the bacterial isolates. KMA and AE performed experiments. KMA and AE analyzed the results and established the tables and figures. KMA and AE wrote the manuscript. FS and TEB revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by Research Ethics Committee of the Faculty of Pharmacy, Tanta University, Egypt, (Research Ethics Committee Code: TP / RE /12-21-M-002).
- Sonbol F, El-Banna T, Elgaml A, Aboelsuod K. Impact of Quorum sensing system on Virulence Factors Production in Pseudomonas aeruginosa. Research. J Pure Appl Microbiol. 2022;16(2):1226-1238.
Crossref - Zhong L, Ravichandran V, Zhang N, et al. Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions. Int J Mol Sci. 2020;21(6):2190.
Crossref - Eladawy M, El-Mowafy M, El-Sokkary MMA, Barwa R. Antimicrobial resistance and virulence characteristics in ERIC-PCR typed biofilm forming isolates of P. aeruginosa. Microb Pathog. 2021;158:105042.
Crossref - Castanheira M, Doyle TB, Deshpande LM, Mendes RE, Sader HS. Activity of ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam against carbapenemase-negative carbapenem-resistant Enterobacterales isolates from US hospitals. Int J Antimicrob Agents. 2021;58(5):106439.
Crossref - Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Khalil MAF, Elgaml A, El-Mowafy M. Emergence of Multidrug-Resistant New Delhi Metallo-β-Lactamase-1-Producing Klebsiella pneumoniae in Egypt. Microbial Drug Resistance (Larchmont, NY). 2017;23(4):480-487.
Crossref - Hager R, Ahmed S, Salam A, Abo N, Magd E. Emergence of carbapenem resistant enterobacteriacae coharboring New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC) isolated from a tertiary hospital in Egypt. Ijambr. 2019;7:86-94.
- Ahmed El-Domany R, El-Banna T, Sonbol F, Abu-Sayedahmed SH. Co-existence of NDM-1 and OXA-48 genes in Carbapenem Resistant Klebsiella pneumoniae clinical isolates in Kafrelsheikh, Egypt. Afr Health Sci. 2021;21(2):489-496.
Crossref - Hoang CQ, Nguyen HD, Vu HQ, et al. Emergence of New Delhi Metallo-Beta-Lactamase (NDM) and Klebsiella pneumoniae Carbapenemase (KPC) Production by Escherichia coli and Klebsiella pneumoniae in Southern Vietnam and Appropriate Methods of Detection: A Cross-Sectional Study. BioMed Res Int. 2019;9757625.
Crossref - El-Mahdy R, El-Kannishy G. Virulence Factors Of Carbapenem-Resistant Pseudomonas aeruginosa In Hospital-Acquired Infections In Mansoura, Egypt. Infect Drug Resist. 2019;12:3455-3461.
Crossref - El-Domany RA, Emara M, El-Magd MA, Moustafa WH, Abdeltwab NM. Emergence of Imipenem-Resistant Pseudomonas aeruginosa Clinical Isolates from Egypt Coharboring VIM and IMP Carbapenemases. Microb Drug Resist. 2017;23(6):682-686.
Crossref - Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-458.
Crossref - Shaaban M, Al-Qahtani A, Al-Ahdal M, Barwa R. Molecular characterization of resistance mechanisms in Pseudomonas aeruginosa isolates resistant to carbapenems. J Infect Dev Ctries. 2018;11(12):935-943.
Crossref - Zafer MM, Amin M, El Mahallawy H, Ashour MSE-D, Al Agamy M. First report of NDM-1-producing Pseudomonas aeruginosa in Egypt. Int J Infect Dis. 2014;29:80-81.
Crossref - Bassetti M, Giacobbe DR, Giamarellou H, et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin Microbiol Infect. 2018;24(2):133-144.
Crossref - Liu X, Wu Y, Zhu Y, et al. Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerg Microbes Infect. 2022;11(1):648-661.
Crossref - Smelikova E, Tkadlec J, Krutova M. How to: screening for mcr-mediated resistance to colistin. Clin Microbiol Infect. 2022;28(1):43-50.
Crossref - Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-168.
Crossref - Zhang H, Yu F, Lu X, et al. Rapid Detection of MCR-Mediated Colistin Resistance in Escherichia coli. Microbiol Spectrm. 2022;10(3):e0092022.
Crossref - Rossi Goncalves I, Dantas RCC, Ferreira ML, Batistao D, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48(2):211-217.
Crossref - Collee JG RSM, Wan B. Tests for the identification of bacteria. In J.G. Collee, A.G. Fraser, B.P. Marmion, and A. Simmons (eds.). Mackie and McCartney Practical Medical Microbiology 14th ed Churchill Livingstone, Edinburgh ed 1996:131-150.
- Uwizeyimana JD, Kim D, Lee H, Byun JH, Yong D. Determination of Colistin Resistance by Simple Disk Diffusion Test Using Modified Mueller-Hinton Agar. Ann Lab Med. 2020;40(4):306-311.
Crossref - Wang JT, Wu UI, Lauderdale TL, et al. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PloS One. 2015;10(3):e0121668.
Crossref - Pollack M. The Virulence of Pseudomonas aeruginosa. Rev Infect Dis. 1984;6(Suppl 3):S617-S626.
Crossref - Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3(4):189-193.
- Goldstein C, Lee MD, Sanchez S, et al. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother. 2001;45(3):723-726.
Crossref - Tsakris A, Themeli-Digalaki K, Poulou A, et al. Comparative evaluation of combined-disk tests using different boronic acid compounds for detection of Klebsiella pneumoniae carbapenemase-producing enterobacteriaceae clinical isolates. J Clin Microbiol. 2011;49(8):2804-2809.
Crossref - Emara M, Abd-Elmonsef M, Abo Elnasr L, Elfeky A. Study of mcr-1 gene-mediated colistin resistance in Gram-negative isolates in Egypt. J Egypt J Med Microbiol. 2019;28(3):9-16.
Crossref - Jeon BC, Jeong SH, Bae IK, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in korea. J Clin Microbiol. 2005;43(5):2241-2245.
Crossref - Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011;49(2):718-721.
Crossref - Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J. 2012;3(1):19-24.
Crossref - Venkatachalam I, Teo J, Balm MN, Fisher DA, Jureen R, Lin RT. Klebsiella pneumoniae Carbapenemase-producing enterobacteria in hospital, Singapore. Emerg Infect Dis. 2012;18(8):1381-1383.
Crossref - Xia Y, Liang Z, Su X, Xiong Y. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32(4):270-275.
Crossref - Sechi LA, Zanetti S, Dupré I, Delogu G, Fadda G. Enterobacterial repetitive intergenic consensus sequences as molecular targets for typing of Mycobacterium tuberculosis strains. J Clin Microbiol. 1998;36(1):128-132.
Crossref - Heras J, Domínguez C, Mata E, et al. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015;16:270.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Migiyama Y, Yanagihara K, Kaku N, et al. Pseudomonas aeruginosa Bacteremia among Immunocompetent and Immunocompromised Patients: Relation to Initial Antibiotic Therapy and Survival. Jpn J Infect Dis. 2016;69(2):91-96.
Crossref - Cho HH, Kwon KC, Kim S, Park Y, Koo SH. Association between Biofilm Formation and Antimicrobial Resistance in Carbapenem-Resistant Pseudomonas Aeruginosa. Ann Clin Lab Sci. 2018;48(3):363-368.
- Laupland KB, Parkins MD, Church DL, et al. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-beta-lactamase (MBL)-producing strains. J Infect Dis. 2005;192(9):1606-1612.
Crossref - Moosavian M, Rahimzadeh M. Molecular detection of metallo-β-lactamase genes, bla IMP-1, bla VIM-2 and bla SPM-1 in imipenem resistant Pseudomonas aeruginosa isolated from clinical specimens in teaching hospitals of Ahvaz, Iran. Iran J Microbiol. 2015;7(1):2-6.
- Falahat S, Shojapour M, Sadeghi A. Detection of KPC Carbapenemase in Pseudomonas aeruginosa Isolated From Clinical Samples Using Modified Hodge Test and Boronic Acid Phenotypic Methods and Their Comparison With the Polymerase Chain Reaction. Jundishapur J Microbiol. 2016;9(9):e27249-e27249.
Crossref - Othman HB, Halim RMA, Abdul-Wahab HEE, Atta HA, Shaaban O. Pseudomonas aeruginosa – Modified Hodge Test (PAE-MHT) and ChromID Carba Agar for Detection of Carbapenemase Producing Pseudomonas Aeruginosa Recovered from Clinical Specimens. Open Access Maced J Med Sci. 2018;6(12):2283-2289.
Crossref - Pasteran F, Veliz O, Faccone D, et al. A simple test for the detection of KPC and metallo-β-lactamase carbapenemase-producing Pseudomonas aeruginosa isolates with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect. 2011;17(9):1438-1441.
Crossref - Neyestanaki DK, Mirsalehian A, Rezagholizadeh F, Jabalameli F, Taherikalani M, Emaneini M. Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns. 2014;40(8):1556-1561.
Crossref - Kosykowska E, Szymanek-Majchrzak K, Walter de Walthoffen S, et al. Molecular analysis of carbapenem-resistant strains of Pseudomonas aeruginosa isolated from patients hospitalized in various transplantation wards between 2008 and 2011. Transplant Proc. 2014;46(8):2576-2578.
Crossref - Robledo IE, Aquino EE, Vázquez GJ. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother. 2011;55(6):2968-2970.
Crossref - Ali DO, Nagla MMA. Molecular Detection of & lt;em>bla</em> OXA-48 Gene Encoding Carbapenem Resistance Pseudomonas aeruginosa Clinical Isolates from Khartoum State Hospitals, Sudan. medRxiv. 2020:06.22.20137034.
Crossref - Gai Z, Samodelov SL, Kullak-Ublick GA, Visentin M. Molecular Mechanisms of Colistin-Induced Nephrotoxicity. Molecules. 2019;24(3).
Crossref - Sader HS, Flamm RK, Carvalhaes CG, Castanheira M. Antimicrobial Susceptibility of Pseudomonas aeruginosa to Ceftazidime-Avibactam, Ceftolozane-Tazobactam, Piperacillin-Tazobactam, and Meropenem Stratified by U.S. Census Divisions: Results from the 2017 INFORM Program. Antimicrob Agents Chemother. 2018;62(12).
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.