ISSN: 0973-7510
E-ISSN: 2581-690X
Several studies have suggested that irrespective of the mode of action of antibiotics, induction of oxidative stress also contributes to their bactericidal actions. When antioxidants are supplemented with antibiotics, the bactericidal effect of the antibiotic are reduced. Thus, in this study, sensitivity of norfloxacin was analysed in the presence of antioxidants like glutathione and ascorbic acids, then the influence of multivitamins in the sensitivity was also analysed. The effect was studied though disc diffusion, MIC (minimal inhibitory concentration) and DNA damage assays against the model organisms – Pseudomonas aeruginosa and Klebsiella pneumoniae. This study suggested that the antioxidant and vitamin solution were having influence over sensitivity towards the antibiotics whose MIC was determined at highest concentration with reduced zone of inhibition and DNA damage. Hence, these results lead to a hypothetical claim for the involvement of multivitamins / antioxidants to have antibacterial resistance development.
Pseudomonas aeruginosa, Klebsiella pneumoniae,Norfloxacin, Oxidative stress, multivitamin.
The advent of antibiotics has proven critical to modern medicine as several clinical procedures would be practically impossible without them (due to the risk of infection). The ground-breaking discovery of penicillin in 1928, sulfonamides in the 1930s, the ‘antibiotic revolution’ from the 1940s to the 1960s and the following decades of successful drug-potentiating antibiotic modifications have contributed significantly to the clinical arma-mentarium against bacterial infections1,2. Recent reviews have described the prevalence and contribution of each resistance mechanism to each class of antibiotics in detail3-6.
Equally critical is utilization of antibiotics and the knowledge about susceptibility of bacteria to a given antibiotic. Sensitivity or resistance to an antibacterial agent largely depends on the type of bacteria treated. The major classes of available antibiotics function via the selective inhibition of bacterial cell wall synthesis7, protein synthesis8, nucleic acid synthesis/ replication9 or the alteration of bacterial cell membrane functions10,11. Selective cell wall, differences in ribosome, nucleic acid biosynthesis pathway and cell membrane structure made them to be specific in action towards the target bacterial cells. Several studies now suggest that cell death by all major classes of bactericidal antibiotics, are not only because of action on specific target but also, by generation of oxidative stress. Dwyer et al12 reported the bactericidal antibiotics to induce the production of hydroxyl radical via the depletion of NADH in the Tricarboxylic Acid (TCA) cycle and electron transport system (ETC). They found norfloxacin, ampicillin and kanamycin to induce a surge in NADH consumption leading to increased production of superoxide radicals which promote the destabilization of iron-sulfur clusters in the ETC complexes resulting in the leaching of iron and generation of hydroxyl radicals13. Thus produced hydroxyl radicals cause extensive damage to biological macromolecules of bacteria resulting in cell death12. From the foregoing discourse it can be inferred that since the bactericidal actions of antibiotics involves their ability to induce oxidative stress and consequent pro-oxidant-mediated damage, its bactericidal actions can be abated by anti-oxidants which scavenge the produced free radicals. Several research works dealing with bactericidal effect of antibiotics in the presence of antioxidants for instance glutathione and ascorbic acid have proved the protective role of these antioxidants against the actions of antibiotics on Escherichia coli14 with a significant increase in the minimum inhibitory concentrations (MICs) of ciprofloxacin14 and streptomycin15 in the presence of antioxidants. Norfloxacin-induced mutagenesis was found to be decreasing in Salmonella typhimurium in the presence of vitamins C and E16. This research investigation was done to investigate the synergistic effect and DNA-protective activity of multivitamins on the sensitivity of P. aeruginosa and K. pneumoniae against norfloxacin (a wide-spectrum, 2nd generation quinolone). The synergistic effect of multivitamins on the bactericidal actions of antibiotic is yet to be studied.
Bacterial Strains
Pseudomonas aeruginosa (ATCC 27853) and Klebsiella pneumoniae (ATCC 13863) were obtained from the American Type Culture Collection (ATCC) and used in this research investigation. The purity and viability of the cultures were tested using several selective media. Cetrimide Agar was used for P. aeruginosa while MacConkey Agar was used for the lactose fermenter, K. pneumoniae.
Culture Media, Antimicrobial Agents and Multivitamins
The Kirby-Bauer antibiotic susceptibility testing17 was carried out on Mueller-Hinton Agar (MHA; HiMedia M173) while the Minimum Bactericidal Concentration (MBC) plating was done on Nutrient Agar plates (HiMedia M001). Minimum Inhibitory Concentration (MIC) testing via tube dilution method was carried out in Mueller-Hinton Broth (HiMedia M391). The antibacterial agent used for sensitivity testing was norfloxacin (Sigma-Aldrich) and the antioxidants used were Glutathione (Hi-media), L-ascorbic (Hi-Media) and multivitamin solution/ 5mL (Lysine (L-lysine HCl – 100mg), Vitamin A – 1000 IU, Vitamin D – 200 IU, Vitamin E – 5 IU, Vitamin B1 – 0.5mg, Vitamin B2 – 0.5 mg, Nicotinamide 6mg, Vitamin C 15 mg, pantothenic acid – 2mg, Vitamin B12). The brand name of the multivitamin used in this experiment is anonymized for confidentiality.
Antibiotic Sensitivity: Synergistic Effects of Antioxidants
Disc diffusion and MIC
The prepared overnight cultures were lawn onto the Mueller Hinton Agar (MHA) using sterile (autoclaved) cotton swabs to spread them evenly across the entire surface of the agar plate and then allowed to dry. Four sterile (autoclaved) Whatman no. 1 filter paper (6 mm diameter), containing 40, 200, 400, and 2000 ng/ disc of antibiotics were then placed on the MHA. These disks were placed about 24 mm apart to avoid overlapping of the zones of inhibition. In order to determine the rescue effect of antioxidants, 5 ml of 10 mM Glutathione-ascorbic acid and 5 mL of multivitamin solution were aseptically prepared and incorporated into the respective agar preparation. After an overnight incubation of the plates, the zones of complete inhibition were measured and recorded. MIC was determined using the tube dilution method. This was carried out in culture tubes containing 2 mL of broth into which 20µL of overnight cultured bacteria were inoculated. Apart from the negative control, the other nine tubes were supplemented with a doubling increment of antibiotics with concentrations ranging from 25 to 6400 ng. MBC of each culture condition was determined by plating a single streak of the culture on a nutrient agar plate using a sterile loop. These plates were incubated overnight at 37°C and were observed for culture growth.
Evaluation of DNA damage
DNA Fragmentation Assay
Genomic DNA of the treated bacterial cells from the MIC assay were extracted using the Phenol-chloroform method18. Following spectrophotometric quantification of the extracted DNA, 40µg of genomic DNA from each assay condition were separated using a gel containing 0.8% agarose in 0.5X TBE buffer. Lambda DNA Hind III digest was used as marker to determine the molecular weight of the resolved fragments. Ethidium bromide was used for the visualization of the generated DNA bands.
Gel Analysis
Analysis of gel image (using Alpha Imager® HP Systems) was carried out using the gel analysis software, TotalLab Quant (TL 100). Image analyses include molecular weight quantification, lane/band detection and quantity calibration.
Antibiotic Sensitivity: Synergistic Effects of Antioxidants
The results of Kirby-Bauer disc diffusion method for antibiotic susceptibility test showed a dose-dependent bacterial growth inhibition in control against both the bacteria, P. aeruginosa and K. pneumoniae. And an appreciable reduction in the zones of inhibition were observed with the case of glutathione, ascorbic acid, glutathione + ascorbic acid and multivitamin supplemented cultures on comparing with the control (Fig. 1 and 2). The reduced bactericidal action of norfloxacin in the presence of glutathione and ascorbic acid can be attributed to their already known ROS-scavenging ability19. These free radical scavengers reduce the lethality of norfloxacin by supplying the need electrons to the generated hydroxyl radicals and preventing their damage to biomolecules.
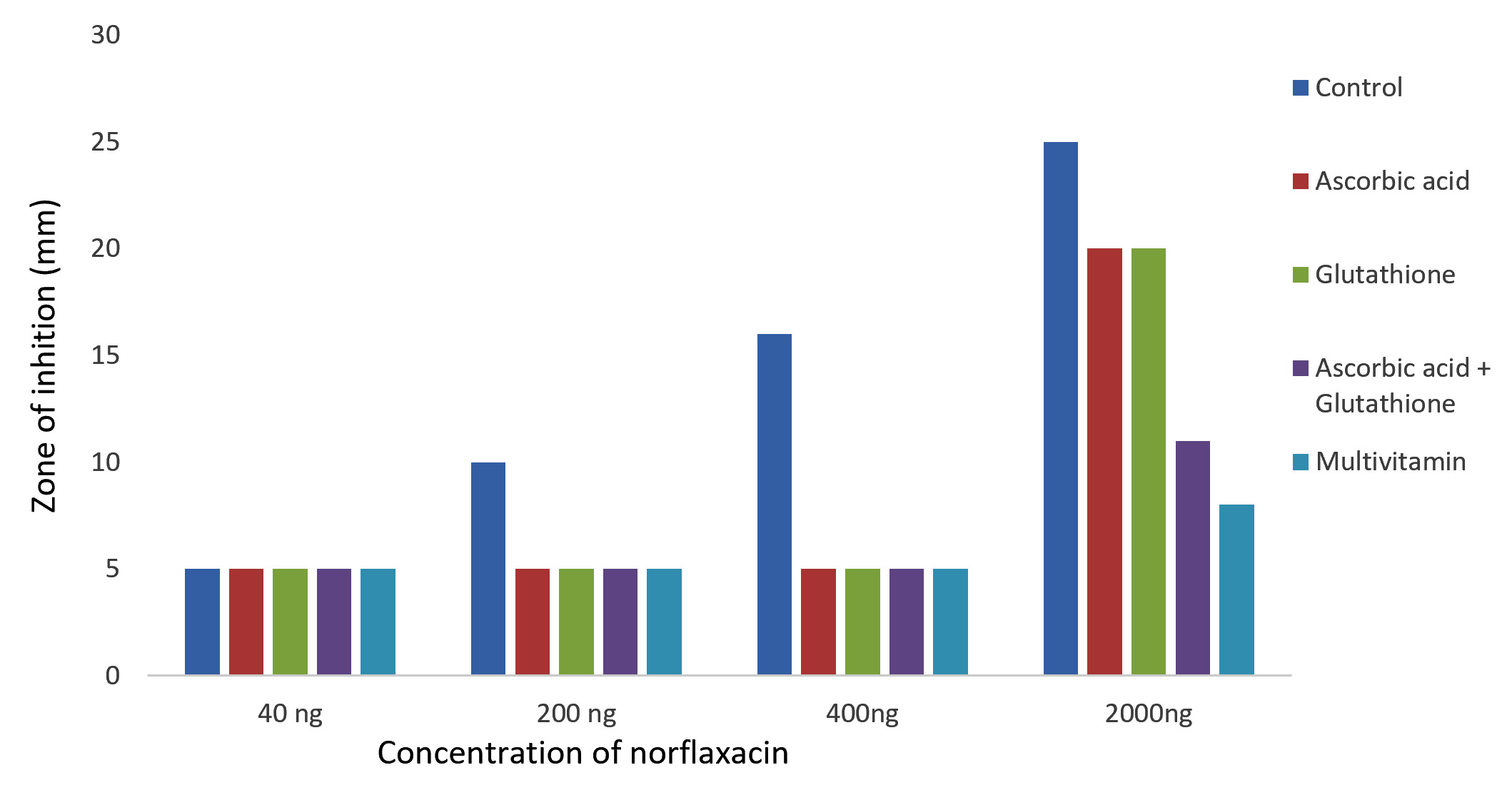
Fig. 1. Effect of antioxidant/multivitamins in bactericidal effect of norfloxacin on Pseudomonas aeruginosa
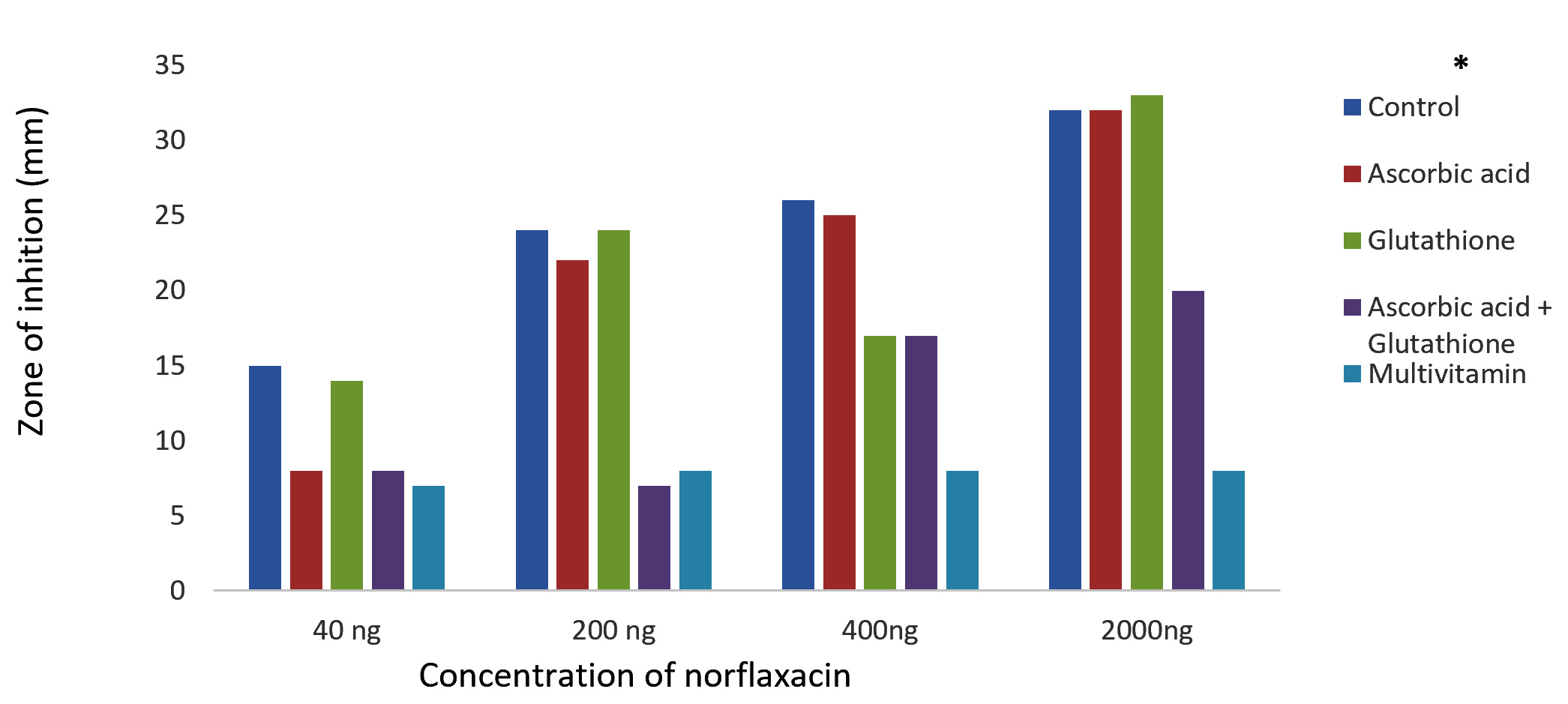
Fig. 2.Effect of antioxidants/multivitamins on the bactericidal effect of norfloxacin on K. pneumoniae
The result for the MIC using the tube dilution method was consistent with the observation in the antibiotic susceptibility tests. As observed from Table 1, the controls for both K. pneumoniae and P. aeruginosa showed growth even at highest concentration of antibiotics used in this study. Even there are evidences for the influence of ascorbic acid and glutathione in resistance against ciprofloxacin and strep-tomycin14,15.
Table (1):
Table showing the MBCs of K. pneumoniae and P. aeruginosa using the tube dilution method. ‘+’ means ‘observable microbial growth’ while ‘0’ means ‘no observable microbial growth’ on culture plate.
| Antibiotic Concentration
(ng) |
0 | 25 | 50 | 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 |
|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | ||||||||||
| Control | + | + | + | + | + | + | 0 | 0 | 0 | 0 |
| Ascorbic Acid
(10 mM) |
+ | + | + | + | + | + | + | + | + | + |
| Glutathione
(10 mM) |
+ | + | + | + | + | + | + | + | + | + |
| Glu/Asc
(5 mM each) |
+ | + | + | + | + | + | + | + | + | + |
| Multivitamin
(500 µL) |
+ | + | + | + | + | + | + | + | + | + |
| K. pneumoniae | ||||||||||
| Control | + | + | + | + | 0 | 0 | 0 | 0 | 0 | 0 |
| Ascorbic Acid
(10 mM) |
+ | + | + | + | + | + | + | + | + | + |
| Glutathione
(10 mM) |
+ | + | + | + | + | + | + | + | + | + |
| Glu/Asc
(5 mM each) |
+ | + | + | + | + | + | + | + | + | + |
| Multivitamin
(500 µL) |
+ | + | + | + | + | + | + | + | + | + |
Evaluation of DNA Damage
DNA damage was caused by norflaxacin in both the bacteria used in this study, where glutathione, ascorbic acid and multi vitamins reduced the impact. DNA extracted from multi-vitamin supplemented K. pneumoniae to retain highest concentration of genomic DNA (i.e. 32.65 and 25.02µg respectively) (Fig. 4), whereas, multivitamin supplemented P. aeruginosa showed a highest concentration of intact DNA (Fig. 3). Several studies have shown the free-radical scavenging activity of antioxidants20,21. Presence of specific antibiotics and other environmental stresses induces adaptive resistance in P. aeruginosa against betalactams, amino-glycosides, polymyxins and fluoroquinolone antibiotics22. Several studies corroborate the fact that bacterial resistance occur at sublethal dose of an antibacterial agent23,24. This study is in concordance with the ‘mutant selection window hypothesis’ which claims that the administration of a drug dose that falls below the minimum inhibitory concentration (MIC) of less susceptible mutant subpopulation and above that of more susceptible strains lead to the enrichment and selection of resistant strains23,24.
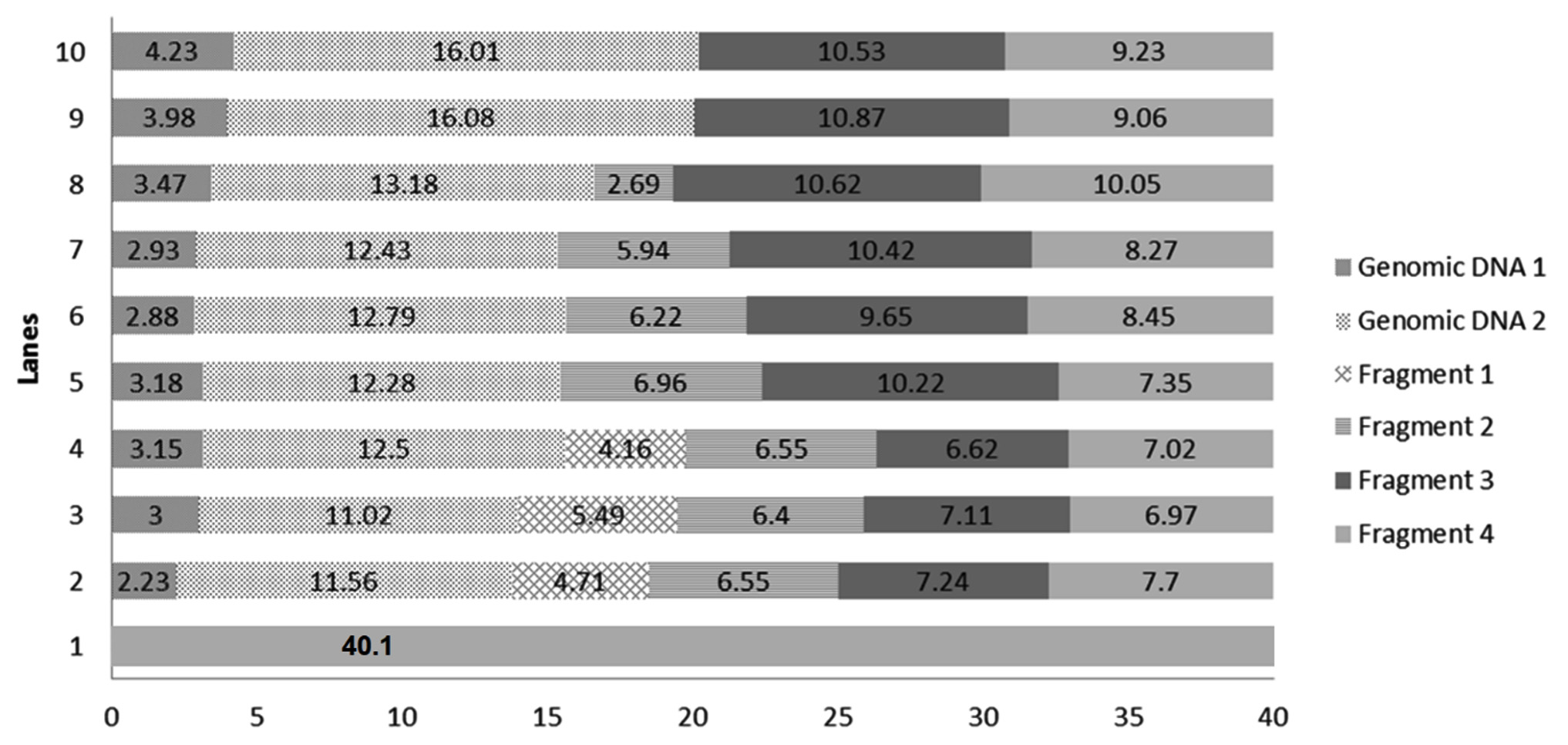 Fig. 3.Chart comparing the DNA quantity (µg) in the generated bands of DNA extracted from selected P. aeruginosa conditions. Genomic DNA 1: quantity of genomic DNA trapped in well, Genomic DNA 2: The first band of DNA, Fragment 1, 2, 3 and 4 are generated DNA fragments. Lane 1: Environmental Control, Lanes 2 – Positive Control with 1600 ng norflaxacin , Lanes 3 and 4 – Ascorbic acid-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 5 and 6 Glutathione-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 7 and 8 – Synergistic study cultures having 800 and 1600 ng norflaxacin respectively, Lanes 9 and 10 – Multivitamin-supplemented cultures having 800 and 1600 ng norflaxacin respectively
Fig. 3.Chart comparing the DNA quantity (µg) in the generated bands of DNA extracted from selected P. aeruginosa conditions. Genomic DNA 1: quantity of genomic DNA trapped in well, Genomic DNA 2: The first band of DNA, Fragment 1, 2, 3 and 4 are generated DNA fragments. Lane 1: Environmental Control, Lanes 2 – Positive Control with 1600 ng norflaxacin , Lanes 3 and 4 – Ascorbic acid-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 5 and 6 Glutathione-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 7 and 8 – Synergistic study cultures having 800 and 1600 ng norflaxacin respectively, Lanes 9 and 10 – Multivitamin-supplemented cultures having 800 and 1600 ng norflaxacin respectively 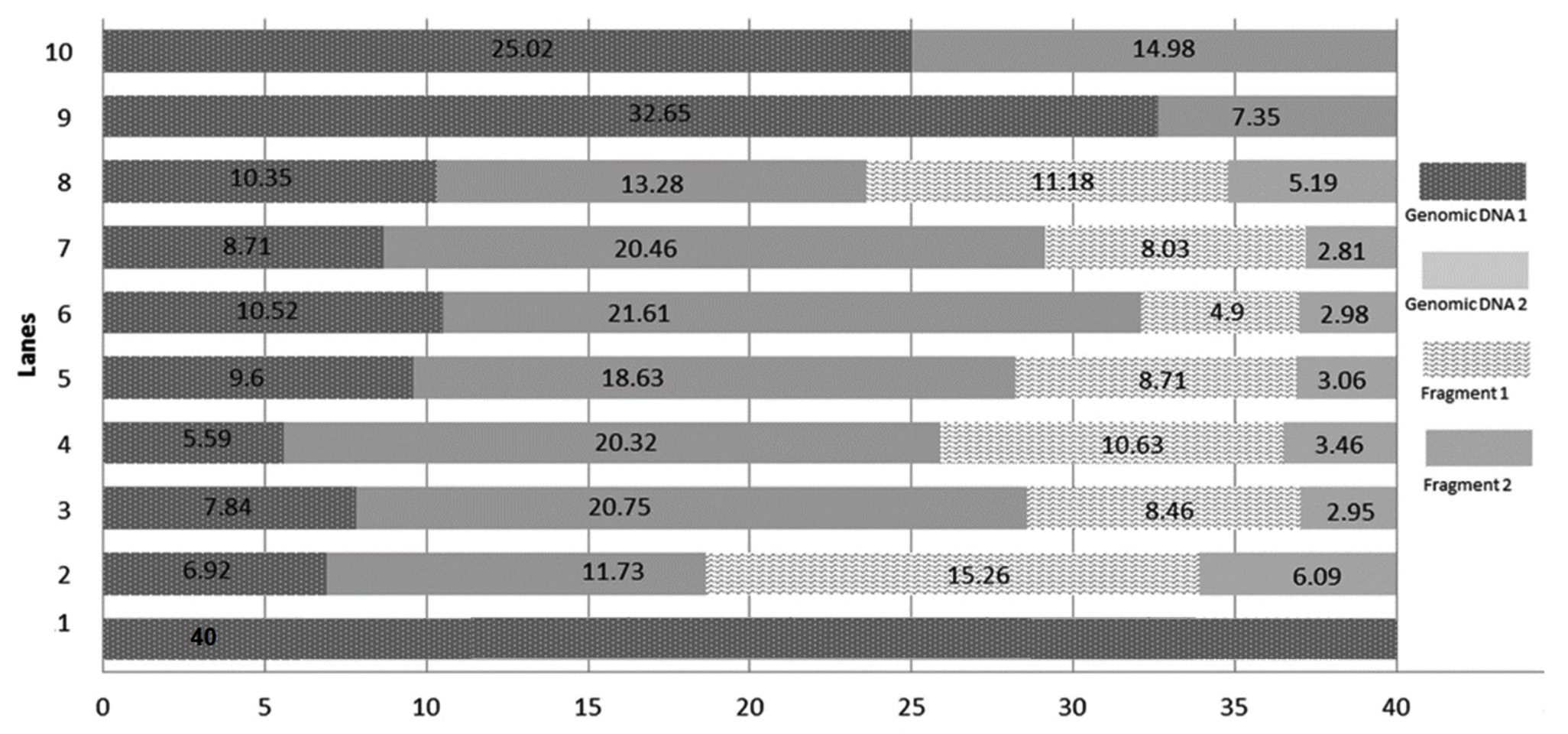 Fig. 4.Chart comparing the DNA quantity (µg) in the generated bands of DNA extracted from selected K. pneumoniae conditions. Genomic DNA 1: quantity of genomic DNA trapped in well, Genomic DNA 2: The first band of DNA observed with molecular weight e” 23.1 kb, Fragment 1 and Fragment 2: generated DNA fragments with molecular weights < 23.1 kb. Lane 1: Environmental Control, Lanes 2 – Positive Control with 1600 ng norflaxacin , Lanes 3 and 4 – Ascorbic acid-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 5 and 6 Glutathione-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 7 and 8 – Synergistic study cultures having 800 and 1600 ng norflaxacin respectively, Lanes 9 and 10 – Multivitamin-supplemented cultures having 800 and 1600 ng norflaxacin respectively
Fig. 4.Chart comparing the DNA quantity (µg) in the generated bands of DNA extracted from selected K. pneumoniae conditions. Genomic DNA 1: quantity of genomic DNA trapped in well, Genomic DNA 2: The first band of DNA observed with molecular weight e” 23.1 kb, Fragment 1 and Fragment 2: generated DNA fragments with molecular weights < 23.1 kb. Lane 1: Environmental Control, Lanes 2 – Positive Control with 1600 ng norflaxacin , Lanes 3 and 4 – Ascorbic acid-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 5 and 6 Glutathione-supplemented cultures having 800 and 1600 ng norflaxacin respectively, Lanes 7 and 8 – Synergistic study cultures having 800 and 1600 ng norflaxacin respectively, Lanes 9 and 10 – Multivitamin-supplemented cultures having 800 and 1600 ng norflaxacin respectively The results of this research investigation further prove the involvement of ROS in the bactericidal actions of norfloxacin against the bacteria chosen in this study. It also suggested that antioxidants (glutathione and ascorbic acid) could reduce the lethality and effectiveness of antibiotics which was evidenced by disc diffusion assay, MIC and DNA damage assay.
Acknowledgements
This work was supported by the Management of MAHSA University for Department of Biomedical Sciences, MAHSA University, Malaysia.
Conflicts Of Interest
The authors declare that there is no conflicts of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Livermore, D.M. Current Epidemiology and Growing Resistance of GramNegative Pathogens. Korean J. Intern. Med., 2012; 27(2): 128–142.
- Moradali, M.F., Ghods, S., Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol., 2017; 7:39.
- Sun, J., Deng, Z., Yan, A. Bacterial multidrug ef ux pumps: Mechanisms, physiology and pharma-cological exploitations. Biochem. Biophys. Res. Commun., 2014; 453: 254–267.
- Hong D.J., Bae I.K., Jang I.H., Jeong S.H., Kang H.K., Lee K. Epidemiology and Characteristics of Metallo-גLactamase-Producing Pseudomonas aeruginosa. Infect. Chemother., 2015; 47(2): 81–97.
- Potron A., Poirel L., Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int. J. Antimicrob. Agents, 2015; 45: 568–585.
- Sun S., Zhou L., Jin K., Jiang H., He Y.W. Quorum sensing systems differentially regulate the production of phenazine1-carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Sci. Rep., 2016; 6: 30352.
- Kong K.F., Schneper L., Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS/ : Acta Pathologica, Micro-biologica, et Immunologica Scandinavica, 2010; 118(1): 1–36.
- Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat., 2010; 13(6): 151–171.
- Fאbrega A., Madurga S., Giralt E., Vila J. Mechanism of action of and resistance to quinolones. Microbial. Biotechnol., 2009; 2(1): 40–61.
- Velkov T., Thompson P.E., Nation R.L., Li J. Structure-Activity Relationships of Polymyxin Antibiotics. J. Med. Chem., 2010; 53(5): 1898–1916.
- Velkov T., Roberts K.D., Nation R.L., Thompson P. E., Li J. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol., 2013; 8(6): 711–724.
- Yeom J., Imlay J.A., Park W. Iron Homeostasis Affects Antibiotic-mediated Cell Death in Pseudomonas species. J. Biol. Chem., 2010; 285(29): 22689–22695.
- Dwyer D.J., Kohanski M.A., Collins J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol., 2009; 12(5): 482–489.
- Goswami M., Mangoli S.H., Jawali N. Involvement of Reactive Oxygen Species in the Action of Ciprofloxacin against Escherichia coli. Anti-microb. Agents Chemother., 2006; 50(3): 949–954.
- Goswami M., Mangoli S.H., Jawali N. Effects of Glutathione and Ascorbic Acid on Streptomycin Sensitivity of Escherichia coli. Antimicrob. Agents Chemother., 2007; 51(3): 1119–1122.
- Alba M.A., Sבnchez R.R., Pיrez N.J.R., Navarrete J. S., Paz R.F., Montoya-Estrada A., Gףmez J.J.H. Comparative study of the antimutagenic properties of vitamins C and E against mutation induced by norfloxacin. BMC Pharmacol., 2008; 8: 2.
- Bauer A.W., Kirby W.M.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Amer. J. Clin. Pathol., 1966; 45(4): 493–496.
- Michael G., Joseph S. Molecular Cloning: A Laboratory Manual. 4th Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2012.
- Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact., 2006; 160(1): 1–40.
- Liu D., Shi J., Ibarra A.C., Kakuda Y., Xue S.J. The scavenging capacity and synergistic effects of lycopene, vitamin E, vitamin C, and beta-carotene mixtures on the DPPH free radical. LWT – Food Sci. Technol., 2008; 41(7): 1344–1349.
- Wang F., Zhao S., Li F., Zhang B., Qu Y., Sun T., Luo T., Li D. Investigation of Antioxidant Interactions between Radix Astragali and Cimicifuga foetida and Identification of Synergistic Antioxidant Compounds. PloS One, 2014; 9(1): e87221.
- Khaledi A., Schniederjans M., Pohl S., Rainer R., Bodenhofer U., Xia, B., Klawonn F., Bruchmann S., Preusse M., Eckweiler D., Dצtsch A., Hהussler S. Transcriptome profiling of antimicrobial resistance in Pseudomonas aeruginosa. Anti-microb. Agents. Chemother., 2016; 60: 4722–4733.
- Ghenghesh K.S., Rahouma A., Tawil K., Zorgani A., Franka, E. Antimicrobial resistance in Libya: 1970-2011. The Libyan Journal of Medicine, 2013; 8: 1–8.
- Drlica K., Hiasa H., Kerns R., Malik M., Mustaev A., Zhao X. Quinolones: Action and Resistance Updated. Curr. Top Med. Chem., 2009; 9(11): 981–998.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


