ISSN: 0973-7510
E-ISSN: 2581-690X
Non-fermenting gram-negative bacteria (NFGNB) frequently exhibit drug resistance. The purpose of this study was to determine the drug resistance pattern among the NFGNB isolates causing respiratory tract infections (RTIs). A retrospective analysis of the antimicrobial susceptibility pattern of non-fermenters causing RTIs over four years (2016- 2019) was done and the change in drug resistance pattern was studied. A total of 653 cases were obtained that included 191 (29.2%) Moraxella catarrhalis, 283 (43.3%) Pseudomonas aeruginosa, and 132 (20.2%) Acinetobacter baumannii, 47 (7.2%) Stenotrophomonas maltophilia isolates. A higher resistance (82.6%) was observed for piperacillin-tazobactam and cefpirome, followed by imipenem (79.5%) and ciprofloxacin (76.5 %) for A. baumannii isolates. A sharp decline in resistance pattern for piperacillin, cefpirome, Imipenem and cefoperazone-sulbactam in 2019 and an increasing resistance to gentamycin and ciprofloxacin were noted. Among P. aeruginosa isolates, 94% aztreonam and 83.4% cefoperazone-sulbactam resistance were detected. There was an increased resistance for cefpirome and piperacillin and a decreased resistance for Imipenem was recorded in 2019. In cases of M. catarrhalis, 22.51% of isolates were resistant to ciprofloxacin, followed by erythromycin (18.32%) and tetracycline (17.80 %). S. maltophilia showed a 100% sensitivity for co-trimoxazole and 2.1% resistance for ciprofloxacin. A constantly changing antibiotic-resistant pattern of non-fermenters compels for a continuous update of drug-resistant trends through a longitudinal surveillance program in different geographical areas.
Acinetobacter baumannii, Pseudomonas aeruginosa, Moraxella catarrhalis, Non-fermenters, Drug-resistant, Respiratory Tract Infection
Non-fermenting gram-negative bacteria (NFGNB) are a diverse group of aerobic, non-spore-forming organisms, lacking the ability to ferment sugars to generate energy for cellular functions.1 NFGNB are widespread saprophytes in the environment and are often considered contaminants of clinical samples, however, the potential pathogenic role is firmly proven by their clinical association with a wide range of diseases.2 Organisms, like Pseudomonas aeruginosa, Acinetobacter baumannii, and Stenotrophomonas maltophilia are often associated with nosocomial infections and patients with immunocompromised status.3,4 NFGNB are known to constitute approximately 16%-21% of all bacterial isolates from a clinical Microbiology laboratory.5,6
NFGNB are becoming a serious challenge to health care settings due to their high resistance to antimicrobial drugs. These bacteria are often intrinsically resistant to various antibiotic groups like macrolides and penicillin. The various resistance mechanisms of NFGNB are mediated by outer-membrane impermeability, alteration of antibiotic target sites, expression of efflux system, production of hydrolyzing enzymes, plasmid, or transposon-mediated resistance.7,8 Antimicrobial drug resistance often increases the treatment difficulties, increases mortality rate, and healthcare expenses.
The antibiotic susceptibility patterns change over time due to an extensive misuse of antibiotics that trigger the selection pressure in microorganisms and enhance the drug resistance phenomenon in different environments.9 The most frequently isolated NFGNB causing nosocomial infections in the intensive care unit (ICU) are P. aeruginosa and A. baumannii.10 In the case of hospital-acquired infections, approximately 10.6% of deaths are caused by multidrug-resistant strains of Acinetobacter spp.11
In this current study, we aimed to determine the drug resistance pattern of non-fermenting gram-negative bacterial isolates, like M. catarrhalis, P. aeruginosa, A. baumannii, and S. maltophilia causing respiratory tract infections in a tertiary care hospital of South Karnataka.
A retrospective study was conducted at the Department of Microbiology in a tertiary care hospital in South Karnataka, over four years from January 2016 to June 2019. The study was approved by the Institutional Ethical Committee (IEC:432/2019). We included all the isolates of M. catarrhalis, P. aeruginosa, A. baumannii, and S. maltophilia that were obtained from the respiratory samples as a part of routine diagnosis.
Microbiological processing of samples
Samples were collected in sterile screw-cap containers and transported to the Microbiology laboratory maintaining the proper cold chain. Upon arrival, samples were inoculated on 5% sheep blood agar and chocolate agar and were also observed for Gram’s staining. All cultures were incubated in 5% CO2 at 37°C for 24 hours. Presumptive identification of all culture-positive cases for M. catarrhalis, P. aeruginosa, A. baumannii, and S. maltophilia was made based on morphology in Gram’s stain and colony characteristics on culture plates. Identification was further confirmed with Matrix-associated Laser Desorption/Ionization- Time of Flight (Vitek MS, bioMerieux Inc., France), and antibiotic susceptibility was performed with the VITEK®2 system (bioMerieux, Inc, Durham, NC).
Data analysis
A structured study proforma was used to document all the relevant demographic and laboratory data of the study subjects. Data were analyzed using SPSS software version 16.0. (IBM). For the categorical variables, descriptive statistical analysis was performed and frequency and percentages were reported. Mean± SD (normal distribution) was used to represent the continuous variables.
A total of 653 NFGNB were included in this study over three and half years, among which 191 (29.2%) were M. catarrhalis, 283 (43.3%) P. aeruginosa, 132 (20.2%) A. baumannii, and 47 (7.2%) S. maltophilia isolates. The 653 isolates were obtained from 420 (64.3%) male patients, and 233 (35.7%) female patients, with a male: female ratio of 1.8:1. The mean age of study cases was 58.8 ±15.6 years, ranging from 1 year to 93 years. The age and gender distribution of the patients infected with NFGNB are given in Table 1.
Table (1):
Age and gender distribution of the patients with Moraxella catarrhalis, Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia respiratory infections.
Moraxella catarrhalis (n=191) |
Acinetobacter baumannii (n= 132) |
Pseudomonas aeruginosa (n=283) |
Stenotrophomonas maltophilia (n= 47) |
Total NFGNB isolates (n=653) |
|
|---|---|---|---|---|---|
Age in years mean ±SD |
58.62 ±15.37 |
59.84 ±16.89 |
57.89 ±15.3 |
61.8 ±15.2 |
58.8 ±15.6 |
>60 years (%) |
109 (57.1) |
79 (59.8) |
138 (48.7) |
26 (55.3) |
258 (43.5) |
30-60 years (%) |
69 (36.1) |
44 (33.4) |
127 (44.9) |
18 (38.3) |
256 (39.2) |
<30 years (%) |
13 (6.8) |
9 (6.8) |
18 (6.4) |
3 (6.4) |
152 (23.3) |
Male (%) |
129 (67.5) |
85 (63.6) |
176 (62.2) |
30 (63.8) |
420 (64.3) |
Female (%) |
62 (32.5) |
47 (36.4) |
107 (37.8) |
17 (36.2) |
216 (35.7) |
Antibiotic susceptibility patterns of Acinetobacter baumannii
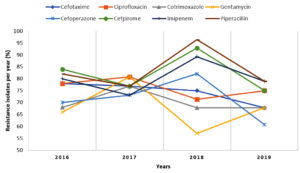
Antibiotic susceptibility was recorded for 132 isolates of A. baumannii (refer to Table 2). Analysis of the susceptibility pattern showed that 109 isolates (82.6%) were resistant to piperacillin-tazobactam and cefpirome, followed by imipenem 105 (79.5%) and ciprofloxacin 101 (76.5 %). The trends of antibiotic susceptibility patterns of A. baumannii isolates are shown in Figure 1. A sharp decline in resistance pattern for piperacillin, cefpirome, Imipenem, and cefoperazone in 2019 and an increasing resistance to gentamycin and ciprofloxacin were noted.
Table (2):
Antibiotic susceptibility of 132 isolates of Acinetobacter baumannii.
Antibiotics |
Sensitive n (%) |
Intermediate n (%) |
Resistant n (%) |
|---|---|---|---|
Cefotaxime |
11 (8.3) |
22 (16.7) |
99 (75) |
Ciprofloxacin |
31 (23.5) |
– |
101 (76.5) |
Cotrimoxazole |
40 (30.3) |
– |
92 (69.7) |
Gentamicin |
42 (31.8) |
1 (0.8) |
89 (67.4) |
Cefoperazone-Sulbactam |
37 (28) |
94 (71.2) |
1 (0.8) |
Cefpirome |
23 (17.4) |
– |
109 (82.6) |
Imipenem |
27 (20.5) |
– |
105 (79.5) |
Piperacillin-Tazobactam |
23 (17.4) |
– |
109 (82.6) |
Antibiotic susceptibility patterns of Pseudomonas aeruginosa
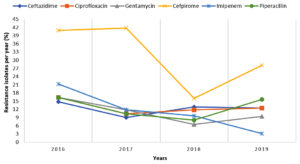
Antibiotic susceptibility of 283 P. aeruginosa isolates is shown in table 3, 94% (266) isolates were found resistant to aztreonam followed by cefoperazone-sulbactam 236 (83.4%) and cefpirome 97 (34.3%). The antibiotic resistance trends over 4 years are given in Figure 2. There was an increased resistance for cefpirome and piperacillin and a decreased resistance for Imipenem was recorded in 2019.
Table (3):
Antibiotic susceptibility of 283 Pseudomonas aeruginosa isolates.
Antibiotics |
Sensitive n (%) |
Intermediate n (%) |
Resistant n (%) |
|---|---|---|---|
Amikacin |
262 (92.6) |
– |
21 (7.4) |
Ceftazidime |
247 (87.3) |
– |
36 (12.7) |
Ciprofloxacin |
244 (86.2) |
1 (0.4) |
38 (13.4) |
Gentamicin |
248 (87.6) |
– |
35 (12.4) |
Aztreonam |
16 (5.7) |
1 (0.4) |
266 (94) |
Cefoperazone-Sulbactam |
46 (16.3) |
1 (0.4) |
236 (83.4) |
Cefpirome |
173 (61.1) |
13 (4.6) |
97 (34.3) |
Imipenem |
242 (85.5) |
– |
41 (14.5) |
Piperacillin-Tazobactam |
243 (85.9) |
3 (1.1) |
37 (13.1) |
Antibiotic susceptibility patterns of Moraxella catarrhalis
M. catarrhalis was isolated from 191 cases, among which 43 (22.51%) were resistant to ciprofloxacin, followed by 35 (18.32%) erythromycin and 34 (17.80 %) tetracycline resistant isolates. A detailed description of the antimicrobial susceptibility of the M. catarrhalis isolates is shown in Table 4.
Table (4):
Moraxella catarrhalis antibiotic susceptibility data of 191 isolates.
Antibiotic |
Sensitive n (%) |
Resistant n (%) |
|---|---|---|
Amoxicillin clavulanate |
187 (97.91) |
4 (2.09) |
Cefotaxime |
185 (96.86) |
6 (3.14 |
Cefuroxime |
160 (83.77) |
31 (16.23) |
Ciprofloxacin |
148 (77.49) |
43 (22.51) |
Erythromycin |
156 (81.68) |
35 (18.32) |
Tetracycline |
157 (82.20) |
34 (17.80) |
Antibiotic susceptibility patterns of Stenotrophomonas maltophilia
Antimicrobial susceptibility of 47 (7.2 %) isolates of S. maltophilia was recorded (refer to Table 5), which shows 100% sensitivity for co-trimoxazole and 2.1% resistance for ciprofloxacin.
Table (5):
Stenotrophomonas maltophilia antibiotic susceptibility data of 47 isolates.
Antibiotic |
Sensitive n (%) |
Resistant n (%) |
|---|---|---|
Ciprofloxacin |
46 (97.9) |
1 (2.1) |
Co-trimoxazole |
47 (100) |
– |
Antibiotics are a potent weapon to fight against bacterial infections, but the rapid emergence of drug-resistant bacteria becoming a threat to healthcare systems. Monitoring the trends of antimicrobial susceptibility in different geographical and hospital settings has prime significance to lead to successful treatment outcomes.
Acinetobacter baumannii is one of the major pathogens associated with nosocomial infection, shows a high level of drug resistance to the available antibiotics, and is often responsible for the outbreak of infections in healthcare settings.12 Studies have reported a 28.2-43.5% isolation rate of the Acinetobacter spp. from various respiratory specimens.13-15 In the present study, the isolation rate of Acinetobacter was 29.2%. The majority of these isolates (82.6%) in our study were resistant to piperacillin-tazobactam and cefpirome followed by 79.5% to imipenem and 76.5% to ciprofloxacin. Another study from Punjab recorded 82.9% of ceftazidime resistance, followed by cefoperazone (82.4%), ciprofloxacin (73.4%), gentamicin (70.4%), ampicillin/sulbactam (58%), amikacin (57.9%), whereas lower resistance for imipenem (23.6%), piperacillin/tazobactam (15.1%) and, sulbactam-ceftazidime (18.4%).16 Bhattacharya et al reported 80% ceftazidime, 55.5% amikacin, and 52.17% ciprofloxacin resistance.17 The distinct resistant pattern among the different studies could be influenced by variation in environmental factors of different settings, unrestricted use of antibiotics, and lack of awareness among clinicians about using broad-spectrum antibiotics.
In this present study, a sharp decline in the resistance trend of certain antibiotics like piperacillin, cefpirome, imipenem, and cefoperazone-sulbactam was observed for Acinetobacter spp. in 2019 compared to 2018, whereas an increasing resistant pattern was observed for Gentamycin and ciprofloxacin. A decreasing trend of antibiotic resistance especially to piperacillin-tazobactam, Imipenem, and cefoperazone-sulbactam can be attributed to effective measures taken by the Hospital Infection Control team in this time period, like the controlled use of carbapenems and higher group of antibiotics by clinicians and spreading more awareness about the correct use of antibiotics in our settings.
Gupta et al. showed decreased resistant isolates of Acinetobacter spp. with a significant linear pattern from 2013-2017.18 Another study from China published an increasing trend of imipenem resistance for the A. baumannii isolates from 2004 (13.3%) to 2014 (70.5%) and also a sharp increase in extensively drug-resistant isolates of A. baumannii from 11.1% (2004) to 60.4% (2014) was noted.19
Pseudomonas aeruginosa is another common isolate in respiratory tract infections, especially in cystic fibrosis and ventilator-associated pneumonia. In the present study, the rate of isolation of P. aeruginosa was found to be 43.3% whereas 39.1% isolation of P. aeruginosa was reported by Sharma et al.3 We observed a high resistance of these isolates against aztreonam (94%), cefoperazone-sulbactam (83.4%), and cefpirome (34.3%). Fatima et al. noted resistance for piperacillin/tazobactam and cefepime at 42% and 40% respectively for P. aeruginosa and an increasing resistance for amikacin (35%).20 A study from Peshawar reported 83.10% cefoperazone-sulbactam and 66.20% piperacillin-tazobactam resistance and the highest sensitivity for amikacin (92.86%).21 Joseph et al. reported a decreasing resistance pattern for ciprofloxacin (49% to 33%), meropenem (35% to 19%), ceftazidime (50% to 33%), and imipenem (28% to 14%) in 2012 in comparison to 2007, limited use of ciprofloxacin may justify this decreasing trend.22 In our study, increasing trends of resistance for cefpirome and piperacillin were observed in 2019 compared to 2018, which may be due to the more common use of these antibiotics for Pseudomonas spp. whereas a decreasing trend of Imipenem resistance was observed in our study, because of the controlled use of carbapenems in our hospital.
Moraxella catarrhalis, commonly causing respiratory infections, is intensively studied in the USA and European countries but very little research has been conducted to date in developing countries like India. According to a few recent reports, M. catarrhalis was isolated in 6.45% to 20% of the respiratory samples.23-25 The isolation rate of M. catarrhalis was found to be 20.2% which is similar as compared to the previously reported studies. In our study, 22.5% of isolates were resistant to ciprofloxacin and 17.8% to tetracycline. Gupta et al. from Delhi observed 15.8% of ciprofloxacin and 14.2% tetracycline resistant isolates, which was more or less similar to our findings.26 Another study from India reported 5% of cefuroxime and 15% of amoxicillin-clavulanate resistant isolates, in comparison to 16.23% and 2.09% resistant isolates of M. catarrhalis that were observed respectively, in our study.24 Krishna et al. reported 13.72% of cefotaxime resistant isolates, which was 3.14% in our study, a higher use of these drugs in the healthcare settings may justify it.25 18.32% of M. catarrhalis isolates in the present study were resistant to erythromycin, whereas a study from China reported a higher resistance rate to erythromycin (70.8%).27
S. maltophilia, is an opportunistic pathogen, particularly among immunocompromised and hospitalized patients with a significant fatality rate.28 The isolation rate of S. maltophilia was noted at 7.2% in our study, whereas previous studies reported a 2.6- 5.5% of isolation rate.29,30 S. maltophilia isolates were 100% sensitive to co-trimoxazole but 2.1% were resistant to ciprofloxacin in our settings. Nayyar et al. reported that 8.6% of isolates were resistant to co-trimoxazole and no resistance was observed against levofloxacin.30 Abdel-Aziz N et al. reported a 100% sensitivity for co-trimoxazole, whereas Azimi A et al. from Iran observed 10.9% resistance to ciprofloxacin.31,32
The alarming increasing rate of drug resistance isolates due to mutations is a critical global health challenge, which requires a regulated use of antibiotics and raising awareness among the community and healthcare personnel about the appropriate use of antibiotics. The retrospective nature of the present study is one of the major limitation. Moreover, we are unable to determine the antibiotic resistance trends of Moraxella catarrhalis, due to a change in CLSI guidelines for the susceptibility range.
A constantly changing antibiotic-resistant pattern of non-fermenters observed in the current study demands a continuous update of drug-resistant trends through a longitudinal surveillance program in different geographical areas. This will help the hospital infection control team to develop antimicrobial guidelines and policies to regulate the spread of resistance and effective management of the infections.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
KC conceptualized the study, performed supervision. KMK collected and compiled the data. AB supervised in data collection and data analysis. DH performed data curation. AB and DH wrote the manuscript. KC and PAS reviewed the manuscript. KC revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
The study was approved by the Kasturba Hospital and Kasturba Medical College Institutional Ethical Committee (IEC:432/2019).
- Koneman E, Allen S, Janda W, Schreckenberger P, Winn W. Colour Atlas and textbook of Diagnostic Microbiology. 6th Edition, Lippincott -Raven Publishers, Philadelphia, 2006.
- Gokale SK, Metgud SC. Characterization and antibiotic sensitivity pattern of nonfermenting gram negative bacilli from various clinical samples in a tertiary care hospital, Belgaum. Belgaum J Pharm Biomed Sci. 2012;17(14):1-5. https://www.jpbms.info/index.php?option=com_docman&task=doc_download&gid=438&Itemid=41. Accessed April 12, 2021.
- Sharma S, Pujari S, Sharma AK. Isolation of non-fermenting Gram negative bacteria in respiratory tract infections. IP Int J Med Microbiol Trop Dis. 2020;6(3):184-187.
Crossref - Buzila ER, Nastase EV, Lunca C, Badescu A, Miftode E, Iancu LS. Antibiotic resistance of non-fermenting Gram-negative bacilli isolated at a large Infectious Diseases Hospital in North-Eastern Romania, during an 11-year period. Germs. 2021;11(3):354-362.
Crossref - Juyal D, Negi V, Prakash R, et al. Prevalence of non-fermenting gram-negative bacilli and their in vitro susceptibility pattern in a tertiary care hospital of Uttarakhand: A study from foothills of Himalayas. Saudi J Health Sci. 2013;2(2):108-112.
Crossref - Somily A, Balkhy HH, Enani MA, et al. Antimicrobial resistance trends of non-fermenter Gram negative bacteria in Saudi Arabia: A six-year national study. J. Infect Public Health. 2021;14(9):1144-1150.
Crossref - McGowan Jr JE. Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am J Infect Control. 2006;34(5):S29-37.
Crossref - Higgins CS, Murtough SM, Williamson E, et.al. Resistance to antibiotics and biocides among non-fermenting Gram-negative bacteria. Clin Microbiol Infect. 2001;7(6):308-315.
Crossref - Karam G, Chastre J, Wilcox MH, Vincent JL. Antibiotic strategies in the era of multidrug resistance. Crit Care. 2016;20(1):136.
Crossref - Aly NY, Al-Mousa HH, El Sayed M. Nosocomial infections in a medical-surgical intensive care unit. Med Princ Pract. 2008;17(5):373-377.
Crossref - Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471-3484.
Crossref - Joly-Guillou ML. Clinical impact and pathogenicity of Acinetobacter. Clin Microbiol Infect. 2005;11(11):868-873.
Crossref - Suryawanshi NM, Mangalkar SM, Davane MS. Prevalence of infection by Acinetobacter species and their antibiogram at a tertiary care hospital. Med Int Med J Microbiol. 2017;1(3):43-45. http://medpulse.in/Microbiology/Article/Volume1Issue3/Micro_1_3_3.pdf. Accessed April 10, 2021.
- Vishwanath S, Chawla K, Gopinathan A. Multidrug resistant Gram-negative bacilli in lower respiratory tract infections. Iran J Microbiol. 2013;5(4):323-327.
Crossref - Sharma A, Bawankar S, Kilikdar M. Prevalence and Antibiogram of Acinetobacter Infections: An experience from a Teaching Institute of Rural Setting, in Central India. Int J Curr Microbiol App Sci. 2019;8(1):1674-1678.
Crossref - Sidhu SK, Arora T, Devi P, Malhotra S, Singh K, Kaur M. Frequency and antibiogram of Acinetobacter species isolated from various clinical samples in a tertiary care hospital. Asian Pac J Health Sci. 2017;4(3):263-268.
Crossref - Bhattacharyya S, Bhattacharyya I, Rit K, et al. Antibiogram of Acinetobacter spp. isolated from various clinical specimen in a tertiary care hospital in West Bengal, India. Biomed Res. 2013;24(1):43-46. https://www.alliedacademies.org/articles/antibiogram-of-acinetobacter-spp-isolated-from-various-clinical-specimensin-a-tertiary-care-hospital-in-west-bengal-india.pdf. Accessed March 25, 2021.
- Gupta V, Ye G, Olesky M, Lawrence K, Murray J, Yu K. Trends in resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States: 2013-2017. BMC Infect Dis. 2019;19(1):742.
Crossref - Gao L, Lyu Y, Li Y. Trends in drug resistance of Acinetobacter baumannii over a 10-year period: Nationwide data from the China surveillance of antimicrobial resistance program. Chin Med J. 2017;130(6):659-664.
Crossref - Fatima A, Naqvi SB, Khaliq SA, Perveen S, Jabeen S. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infections. Springer Plus. 2012;1(1):70.
Crossref - Samad A, Ahmed T, Rahim A, Khalil A, Ali I. Antimicrobial susceptibility patterns of clinical isolates of Pseudomonas aeruginosa isolated from patients of respiratory tract infections in a Tertiary Care Hospital, Peshawar. PaK J Med Sci. 2017;33(3):670.
Crossref - Joseph NM, Devi S, Shashikala P, Kanungo R. Changing trend in the antibiotic resistance pattern of Pseudomonas aeruginosa isolated from wound swabs of out-patients and in-patients of a tertiary care hospital. J Clin Diagn Res. 2013;7(10):2170-2172.
Crossref - Shaikh SB, Ahmed Z, Arsalan SA, Shafiq S. Prevalence and resistance pattern of Moraxella catarrhalis in community-acquired lower respiratory tract infections. Infect Drug Resist. 2015;8:263-267.
Crossref - Aiswariya A, Suseela KV, Subi D. Prevalence of Moraxella catarrhalis in patients of lower respiratory tract infection with underlying risk factors. Int J Adv Med. 2017;4(2):442.
Crossref - Krishna S, Sagarika S, Jeer M, et al. Prevalence and Antibiotic Sensitivity Pattern of Moraxella catarrhalis in Patients with Lower Respiratory Tract Infections in a Tertiary Health Care Centre in India. Int J Curr Microbiol App Sci. 2016;5(6):72-78.
Crossref - Gupta N, Arora S, Kundra S. Moraxella catarrhalis as a respiratory pathogen. Indian J Pathol Microbiol. 2011;54(4):769-771. https://www.ijpmonline.org/text.asp?2011/54/4/769/91496. Accessed March 27, 2021.
- Shi W, Wen D, Chen C, et al. β-lactamase production and antibiotic susceptibility pattern of Moraxella catarrhalis isolates collected from two county hospitals in China. BMC Microbiol. 2018;18(1):77.
Crossref - Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25(1):2-41.
Crossref - Malini A, Deepa E, Gokul B, Prasad S. Nonfermenting gram-negative bacilli infections in a tertiary care hospital in Kolar, Karnataka. J Lab Physicians. 2009;1(02):062-066.
Crossref - Nayyar C, Thakur P, Tak V, Saigal K. Stenotrophomonas maltophilia: An Emerging Pathogen in Paediatric Population. J Clin Diagn Res. 2017;11(1):DC08-DC11.
Crossref - Abdel-Aziz N, Morsy MMF, Amin SS, Mohammed KI, Alharbi AE, Alshami I. Threatening problem of Stenotrophomonas maltophilia producing extended-spectrum beta-lactamases: Prevalence and automated antibiotic susceptibility pattern. Clin Microbial. 2013;2(2):1000108.
Crossref - Azimi A, Rezaei F, Yaseri M, Jafari S, Rahbar M, Douraghi M. Emergence of fluoroquinolone resistance and possible mechanisms in clinical isolates of Stenotrophomonas maltophilia from Iran. Sci Rep. 2021;11(1):9582.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.