ISSN: 0973-7510
E-ISSN: 2581-690X
Bacterial endophytes inhabiting medicinal plants are less explored, but are diverse and play crucial roles in regulating growth and development of the host. Metagenomics using Illumina MiSeq platform facilitate whole community level characterization. The present study reports the diversity of bacterial endophytic microflora from the medicinal plant Emilia sonchifolia (Linn.)DC. Metagenomic analysis of medicinal plants leads to the identification of novel organisms or genes which will help the correlative elucidation of plant-microbe interactions. Effective sequences were amplified from 16S rRNA gene V3-V4 variable region. OTU analysis at different taxonomic level clearly catalogues two Phyla viz. Proteobacteria and Firmicutes which belonged to Gammaproteobacteria and Bacilli. In these classes five orders such as Enterobacteriales, Pseudomonadales, Xanthomonadales, Bacillales and Betaproteobacteriales were detected. Among these orders five families were identified in which the most predominant was Enterobacteriaceae and Pseudomonadeaceae while the other three families viz. Xanthomanadaceae, Planococcaceae and Burkholderiaceae were less represented. At genus level very less number of bacteria were identified while a bulk majority remained unclassified. Of the seven identified genus the most prominent one was Pseudomonas followed by Stenotrophomonas, Cronobacter, Lysinibacillus, Pantoea, Kluyvera and Pseudorhodoferax. At species level only two were identified vz. Pseudomonas otitidis and P.geniculate. Alpha diversity analysis using various statistical indices like Simpson and Shannon explains the diversity of microbiome. Next generation sequencing survey of DNA sample extracted from host plant through metagenomic data screening identified different endophytic bacteria which are difficult to grow in culture conditions.
Illumina MiSeq, metagenomics, E.sonchifolia, medicinal plants, endophytic bacteria.
In the past decades, the microbiota associated with plants gained researcher’s attention due to their wide diversity and beneficial applications of plant-microbe interactions. They may be epiphytic or endophytic and their presence can be detected in the rhizospheric or phyllospheric regions (Strobel and Daisy, 2003). Endophytes reside inside the plant tissues asymptomatically and their association can be symbiotic or mutualistic. The endophytic communities of various medicinal plants like Cannabis (McKernan et al., 2018), Aloe vera (Akinsanya et al., 2015), Plectranthus (El-Deeb et al., 2013), Catharanthus, Mentha, and Ocimum (Anjum and Chandra, 2015) were isolated and identified. From these earlier reports it was observed that the interaction between endophytes and host plants play some roles in their medicinal properties.
The understanding of plant-microbe interactions is expanding due to the advancements in next generation sequencing (Lozupone and Knight, 2007). New sequencing platforms like Illumina MiSeq enables tracking of large microbial communities at a faster rate and lower cost (Ram et al., 2011). In the analysis of complex microbial communities like endophytic populations metagenomic sequencing acts as an advanced technique when compared to rDNA sequencing. In this context, Illumina sequencing provides information with fewer errors on biodiversity analysis of microbes (von Mering et al., 2007).
Emilia sonchifolia (Linn.) DC. is a herbaceous medicinal plant used for the treatment of various inflammatory disorders (Shylesh et al., 2000, Chopra et al., 1986, Essien et al., 2009). The multifaceted applications of this medicinal herb has been analysed by many researchers, but the information on the endophytic microbiome and its metataxonomy remain unclear. The present investigation aims to analyse the endophytic bacterial diversity of E. sonchifolia using metagenome analysis through Illumina sequencing platform.
Plant sample collection and surface sterilization
The whole plant of E. Sonchifolia during its flowering season (July-October) was collected from fourteen districts of Kerala (Kasargode 12° 30’ N75° 00 E to Thiruvanathapuram 8° 29’ N76° 59 E), India, pooled and used for DNA extraction after surface sterilization. E. sonchifolia is a herbaceous medicinal plant which is one among the members of ‘Dasapushpa’ (ten flowers of sacred value) in Ayurvedic medicine. It belongs to the family Asteraceae of dicots and possess anti-cancerous, antiinflammatory and analgesic properties (Essien et al., 2009; Shylesh et al.,2000). The whole plant has been used for medicinal purposes and hence the biodiversity analysis was carried out using the entire plant after surface sterilization. Healthy and mature flowering plant was washed thoroughly in running tap water followed by dipping in sterile double distilled water for ten minutes and surface sterilized with 0.1% mercuric chloride for one minute. The surface sterilized material was then rinsed in sterile distilled water and dipped in 70% ethanol for 60 seconds. Further immersion of the plant tissue in distilled water was required to remove the traces of ethanol from the tissue. The effectiveness of the surface sterilization procedure was validated by culturing the final wash into nutrient agar plates. Any bacterial growth on the control plates indicates inadequate surface sterilization.
DNA Isolation and PCR Amplification
DNA was extracted using Purelink genomic DNA extraction and purification kit (Invitrogen, Life Technologies, USA) following the manufacturer’s instruction. After electrophoresis dsDNA concentration was checked by Qubit®4.0 fluorometer. The V3-V4 hyper variable regions from 16S rRNA gene of the purified DNA were amplified using the universal forward primer-5’CCTACGGRRBGCASCAGKVRVGAAT 3’ and reverse primer-5’GGACTACNVGGGTWTCTAA TCC3’.
The amplified DNA was further quantified with Qubit®4.0Fluorometer (Invitrogen, Carlsbad, CA, USA). 30-50ng DNA sample along with Meta VxTM Library preparation kit was used to prepare the amplicons. The PCR amplicons were tagged with adapters for creating indexed libraries. Validation of DNA libraries were done by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified by Qubit®4.0 Flurometer. DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument (Omics Gen Life Sciences Pvt. Ltd.) according to manufactures instructions (Illumina, 2013, Illumina, San Diego, CA, USA). Sequencing was performed using a 2×250 paired-end (PE) configuration. Image analysis and base calling were performed by the MiSeq control software (MCS) with in the MiSeq instrument. The original image data were analysed using bcl2Fastq (V2.17.1.14) and the result was stored in FASTQ format (Ong et al., 2013).
Sequence Data Analysis
The forward and reverse reads were joined and separated on the basis of barcodes. After merging and separation the primers, adapters, barcodes and undetermined bases were removed. This reduces sequencing errors and the analysis software used was Cutadapt [V (1.9.1) V search (1.9.6), Qiime (1.9.1)]. Quality filtering on joined sequences was performed and the sequences without any ambiguous bases, length<200bp, mean quality score≥20 were selected for downstream analysis. As a final step for the preparation of clean data for further analysis chimera sequences were identified by comparing with reference database (RDP Gold data base) using UCHIME algorithm and they were removed.
OTU Analysis
Sequences were grouped into different Operational Taxonomic Units (OTU) based on 97% sequence identity using the clustering Programme V SEARCH (1.9.6) against the Silva 119 database. Taxonomic category at confidence threshold of 0.8 was grouped into different OTU with the help of Ribosomal database programme classifier.
Alpha Diversity Analysis
Alpha diversity analysis was performed to check the microflora biodiversity. The different species in the microbial community and its abundance were calculated through a series of statistical indices like ACE (the number of species), Shannon (Diversity index alpha for the estimation of microbial diversity), Simpson (Quantify biological diversity) and Goods coverage (library coverage of each sample). The analysis software used for the alpha diversity indices was Qiime (1.9.1).
E. sonchifoila, a major medicinal plant of ‘Dasapushpa’ category of Indian traditional medicine, was used for the present analysis. Relative abundance, composition and diversity of endophytic microflora were analysed on Illumina base sequencing platform.
Preliminary Sequencing Data Statistics
Raw read statistics and sequence quality assessments were collected from MiSeq sequencing reporter generated through base calling. Preliminary sequencing data statistics were presented in Table 1. The sequence data has been deposited at NCBI under Sequence Read Archive (SRA) database with accession no. PRJNA542222.
Table (1):
Preliminary data statistics.
Sample Ref No. |
Sequence Format |
Sequence Type |
Read type |
Read size |
Total no of reads |
Total sequence length |
Q 20 |
Q30 |
GC% |
|---|---|---|---|---|---|---|---|---|---|
P_01 |
Fastq |
Illumina MiSeq |
Paired end |
250bp |
155860 |
38965000 |
91.09 |
87.03 |
54.77 |
Sequencing data quality optimization
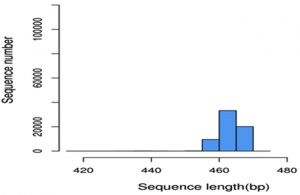
The sequenced data was further processed by analysis software Cutadapt [V (1.9.1)] and Qiime (1.9.1) for the removal of unclassified bacterial reads and the elimination of low complexity reads. Sequence processed was detailed in Table-2, and sequence reads were generated after carry-on overlap splicing through the merging of forward and reverse reads. Alpha diversity analysis require error free sequences and hence it is necessary to remove reads with undesirable length, low quality score, and ambiguous base calls (Ns) (Huse et al., 2008). Reads were trimmed on the above mentioned criteria to reduce the error rate. Out of the 155860 high quality reads obtained, 77930 reads were generated after these filtering processes with average length of 463.45bp (Graph I).
Table (2):
Sequencing data Quality optimization.
Sample |
#PE_reads |
#Nochimera |
Avg Len(bp) |
GC(%) |
|---|---|---|---|---|
P_01 |
77930 |
63453 |
463.45 |
54.48 |
OTU and Taxonomic Composition Analysis
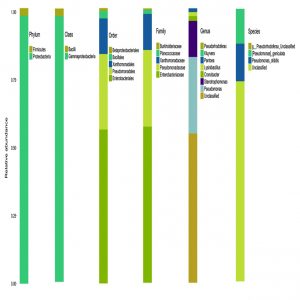
The accurate and high resolution microbiota profiling of endophytic communities can be elucidated through Illumina sequencing platform. All the sequences after filtering and error reduction were classified to obtain information on species and genus. The grouping of sequences relayed on the 97% identity threshold for data statistics and analysis using software Qiime (1.9.1). The number of OTUs of sample was six which is comparatively low with other published metagenomic sequence analysis indicating the host specificity of the endophytic population. The OTU distribution pattern indicated that majority of endophytic bacteria are included under Enterobacteriaceae.
The statistics of the number of species on different taxonomic levels revealed the vast diversity of microbes. The sequence data disclosed phylum Proteobacteria as the predominant taxa followed by Firmicutes and Bacteroidetes. It was interesting to note that the major phylum detected was Proteobacteria but when tried to isolate endophytes on culturable methods the major group was Bacilli from phylum Firmicutes (Urumbil and Anilkumar, 2019).
Few studies have been conducted in medicinal plants for analyzing the biodiversity of bacterial endophytes using Illumina Mi Seq. Illumina based sequencing were employed for the screening of endophytic bacterial diversity in the medicinal plant tree Peony and they reported Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria and acidobacteria as dominat groups of bacterial endophytes(Yang et al., 2017). Panax notoginseng a medicinal plant with antihypertensive, antithrombic and neuroprotective bioactivities were screened for the diversity analysis of bacterial endophytes associated with the plant using the QUIIME Pineline and indicated the presence of Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes as major communities of bacterial endophytes in this plant (Dong et al.,2018). Previous works on endophytes by Sessitsch et al., (2012) reported the dominance of phylum Proteobacteria in the endophytic community. In accordance with the results of Romero et al., (2014), in the present analysis also threw light on the occurrence of Gammaproteobacteria comprising Enterobacterials and Pseudomonadales as key stone taxon.
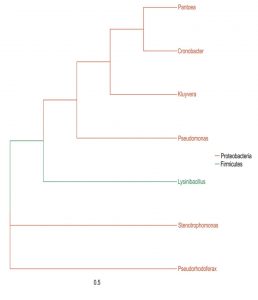
The endophytic bacteria detected from the medicinal plant E.sonchifolia were classified under the orders Enterobacteriales, Psuedomonadales, Xanthomonadales, Bacillales and Betaproteobacteriales. This clearly explained the diversity of endophytic microbiota and similar results were obtained in Poplar trees by Moore et al., (2006). The most dominant genus reported in the present study is Pseudomonas. The other dominant genera were Stenotrophomonas, Cronobactor, Lysinibacillus and Pantoea. Liu et al. (2016 ) reported the dominance of the phylum actinobacteria, proteobacteria and firmicutes in the diversity stydy of endophytic bacterial populations from Ferula songorica. Several members of Enterobacteriales, Psuedomonadales, Bacillales and other orders of bacteria identified in the present study were also reported by various researchers with special mention to the endophyte-host interactive growth promotion (Brigido et al., 2019). These endophytes were found to be effective in preventing the development of disease causing pathogenic fungus in Agave tequilana (Martinez-Rodriguez et al., 2014) and they check the development of wilt disease in Pine Trees (Proença et al., 2017). Role of endophytes in helping host plants for existing in adverse environmental conditions were analysed and the Bacillaceae and Enterobactereaceae were detected as keystone taxa from plants growing in extreme environmental conditions.
A wide variety of bioactive compounds were produced by endophytic bacteria that included the class Gamaproteobacteria and Bacilli points to the significance of studying these categories of bacteria as endophytes (Pimentel et al.,2011 and Gouda et al., 2016). Studies on the medicinal plant Latana camara showed the presence of Bacillus as a prominent member in the endophytic bacterial diversity (Janardhan et al., 2012). Zam et al., (2019) recently reported the occurrence of the genus Bacillus as major endophytic bacteria from 8-10 medicinal plants.
The endophytic bacterial classes present in this study were found to have different levels of interactions with the host plant and they can have wide range of applications. The complete genome sequencing analysis of Enterobactor sp. spotted a number of genes which act as key factors for the dualistic life cycle as soil bacterium and endophyte (Andrés-Barraoet al., 2017). These results support our study on the occurrence of Enterobacteriales as endophytes was common and even their genome analysis proves this factor.
The microbial composition of the sample at different taxonomic level (Phylum, Class, Order, Families, and Genus) was plotted in stalked bar plots (Fig. 1). Phylogenetic tree of the genus constructed and infers approximately maximum likelihood from alignments of the major OTU sequences (Fig 2).
Alpha diversity analysis
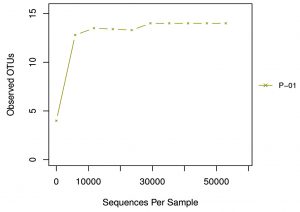
Alpha diversity is mainly used to reflect the species diversity in a single sample through a series of statistical indices like ACE, Chao I, Shannon, Simpson and Good’s coverage using the software Qiime (1.9.1) (Table 3). The rarefaction curve is a useful tool for the species composition characterization in a sample and it predicts the abundance of species in it. It determines whether the sample size is sufficient to estimate the species abundance in biodiversity and community surveys. The alpha diversity and rarefaction analysis (Graph 2) focus on the OTU at 97% similarity and points that bacterial communities were not completely revealed and it was diverse with higher taxonomic richness.
Table (3):
Alpha diversity through a series of statistical indices.
sample |
ace |
chao1 |
shannon |
Simpson |
goods_coverage |
|---|---|---|---|---|---|
P-01 |
14 |
14 |
2.221 |
0.69 |
1 |
Graph 2. Rarefaction curve: The X axis is the valid sequences extracted, and Y axis is the number of OTUs. The number of OUTs increases with the increase of extracted sequence count until reaching a plateau, which indicates the number of detected OTUs will not increase with the amount of extracted sequences and reflects the reasonable sequence depth.
Microflora biodiversity analysis along with metagenome studies on Illumina MiSeq platform enables the proper identification of bacterial population. According to Shi et al.,(2014) endophytes are symbiotic microorganisms and their genetic diversity within the host depends on various other parameters like host genotype, environmental conditions, age of host etc, and was studied in detail on cotton plats by Adams and Kloepper (2002). They also mentioned that the endophytic bacterial diversity studied so far relayed on the culturable communities which is almost <1% of the bacterial endophytic species present. Identity and cellular interactions of these endophytic microbes can be deciphered through metagenomic analysis both in the case of culturable and unculturable bacterial communities (Dinsdale et al., 2008). So this particular study helps for the further identification of some specific endophytic candidates which can contribute to the medicinal properties of this particular host plant E. sonchifolia.
Multidisciplinary research approaches were required for elucidating the beneficial aspects of plant microbe interactions. Metagenomic data analysis is an inevitable complementary information illustrating complex network of factors controlling endophytic colonization and its association with the host. The present study revealed an average read length of 463.45bp from the V3-V4 hypervariable region of 16S rRNA sequences. OTU analysis at different taxonomic level clearly catalogues two Phyla viz. Proteobacteria and Firmicutes which belonged to Gammaproteobacteria and Bacilli. In these classes five orders such as Enterobacteriales, Pseudomonadales, Xanthomonadales, Bacillales and Betaproteobacteriales were detected. Among these orders five families were identified in which the most predominant was Enterobacteriaceae and Pseudomonadeaceae while the other three families viz. Xanthomanadaceae, Planococcaceae and Burkholderiaceae were less represented. At genus level very less number of bacteria were identified while a bulk majority remained unclassified. Of the seven identified genus the most prominent one was Pseudomonas followed by Stenotrophomonas, Cronobacter, Lysinibacillus, Pantoea, Kluyvera and Pseudorhodoferax. At species level only two were identified vz. Pseudomonas otitidis and P. geniculate. Comprehensive knowledge of both culturable and non-culturable endophytes from medicinal plants can unveil the presence of novel compounds and the genes associated with them. In future this can be used as a platform for the development of new drugs. Whole Metagenome data analysis of these endophytes is in progress in our lab and can help in the identification of genes with wide range of applications in biotransformation process.
ACKNOWLEDGMENTS
The first author gratefully acknowledges the financial support and leave under FDP by the University Grants Commission (UGC), New Delhi, Government of India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHOR’S CONTRIBUTION
Both authors have made a substantial direct and intellectual contribution to the work and approved for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All data sets generated during this study are included in the manuscript.
- Adams PD and Kloepper JW. Effect of host genotype on indigenous bacterial endophytes of Cotton (Gossypium hirsutum L.), Plant Soil, 2002; 240: 181-189.
Crossref - Akinsanya MA, Goh JK, Lim SP and Ting ASY. Metagenomic study of endophytic bacteria in Aloe vera using next generation technology, Genomics Data, 2015; 6: 159-163.
Crossref - Andres-Barrao C, Lafi FF, Alam I, de Zelicourt A, Eida AA, Bokhari A, Alzubaidy H, Bajic VB, Hirt H and Saad MM. Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium, Front. Microbiol., 2017; 8(2023): 1-21.
Crossref - Anjum N and Chandra R. Endophytic bacteria: Optimization of isolation procedure from various medicinal plants and their preliminary characterization, Asian J. Pharmaceut. Clinical Res., 2015; 8(4): 233-238.
- Brigido C, Singh S, Menendez E, Tavares MJ, Glick BR, Felix MR, Oliveira S and Carvalho M. Diversity and functionality of cultivable endophytic bacterial communities in Chickpea plants, Plants, 2019; 8(42).
Crossref - Chopra RN, Nayar SL, Chopra IC. Glossary of Indian medicinal plants (Including the supplement), CSIR, New Delhi, 1986.
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, Mc Daniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Broto BR, Ruan Y, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA and Rohwer F. Functional metagenomic profiling of nine biomes, Nature, 2008; 452: 629-632.
Crossref - Dong L, Cheng R, Xiao L, WeiF, Wei G, Xu J, Wang Y, Guo X, Chen Z, Chen S. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng., Chinese Medicine, 2018; 13(41).
Crossref - El-Deeb B, Fayez K and Gherbawy Y. Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their anti microbial activities, J. Plant Interact., 2013; 8(1): 56-64.
Crossref - Essien GE, Nwidu LL and Nwafor PA. Anti-inflammatory and analgesic potential of methanolic extract of Emilia sonchifolia (Compositae) leaves in rodents, Afr. J. Biomed. Res, 2009; 12(3): 199-207.
- Gouda S, Das G, Sen SK, Shin H and Patra JK, Endophytes: A treasure house of bioactive compounds of medicinal importance, Front. Microbiol., 2016; 7(1538):1-8.
Crossref - Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA and Mitchell L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics, 2008; 4(11): 1-11.
Crossref - Illumina, 16S Metagenomic sequencing Library Preparation Guide.http://support. Illumina.com /downloads/16s_metagenomic_sequencing_library_preparation. ilmn., 2013.
- Janardhan BS and Vijayan K. Types of endophytic bacteria associated with traditional medicinal plant Lantana camara Linn., Pharmacognosy Journal, 4(32): 20-23.
Crossref - Liu YH, Guo JW, Salam N, Li L, Zhang YG, Han J, Mohamad OA and Li WJ. Culturable endophytic bacteria associated with medicinal plant Ferula songorica: molecular phylogeny, distribution and screening for industrially important traits, 3 Biotech, 2016: 6(2): 209.
Crossref - Lozupone CA and Knight R. Global patterns in bacterial diversity, Proc. Natl. Acad. Sci., 2007: 104: 11436-11440.
Crossref - Martinez-Rodriguez JC, Mora-Amutio MD, Plascencia-Correa LA, Audelo-Regalado E, Guardado FR, Hernandez-Sanchez E, Pena-Ramirez YJ, Escalante A, Beltran-Garcia MJ and Ogura T. Cultivable endophytic bacteria from leaf bases of Agave tequilana and their role as plant growth promoters. Brazilian J. Microbiol., 2014; 45(4): 1333-1339.
Crossref - McKernan K, Spangler J, Helbert Y, Lynch RC, Deritt-Lee A, Zhang L, Orphe W, Warner J, Foss T, Hudalla CJ, Silva M and Smith DR. Metagenomic analysis of Cannabis samples; Pathogenic bacteria, toxigenic bacteria, and beneficial microbes grow in culture based yeast and mold tests, F1000 Res., 2016; 5(2471): 1-20.
Crossref - Moore FP, Barac T, Borremans B, Oeyen L, Vangronsveld J, van der Lelie D, Campbell CD and Moore ER. Endophytic bacterial diversity in poplar trees growing on the BTEX-contaminated site: the characterization of isolates with potential to enhance phytoremediation, Syst. Appl. Microbiol., 2006; 29: 439-556.
Crossref - Ong SH, Kukkillaya VU, Wilm A, Lay C, Ho EXP, Low L, Hibberd ML and Nagarajan N. Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16s rRNA amplicon sequences, PLoS ONE, 2013; 8(4): 1-8.
Crossref - Pimentel MR, Molina G, Dionisio AP, Marostica Junior, MR, Pastore GM. The use of endophytes to obtain bioactive compounds and their application in Biotransformation process. Biotechnol, Res.Internatl., 2011: 1-11.
Crossref - Proenca DN, Francisco R, Kublik M, Scholer A, Vestergaard G, Schloter M and Morais PV. The mocrobiome of endophytic, wood colonizing bacteria from Pine trees as affected by Pine wilt disease. Sci. Rep., 2017; 7: 4205.
Crossref - Ram JL, Karim AS, Sendler ED and Kato I. Strategy for microbiome analysis using 16s rRNA gene sequence analysis on the Illumina sequencing platform, Systems Biol. Repro. Med., 2011; 57: 162-170.
Crossref - Romero FM, Maria M and Pieckenstain FL. The communities of Tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analysed by 16S ribosomal RNA gene pyro sequencing, FEMS Microbiol. Lett., 2014; 351: 187-194.
Crossref - Sessitsch A, Hardoim P, Doring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD and Reinhold-Hurek B. Functional characteristics of an endophytic community colonizing rice roots as revealed by metagenomic analysis, MPMI, 2012; 25(1): 28-36.
Crossref - Shi YW, Yang H, Zhang T, Sun J and Lou K. Illumina–based analysis of endophytic bacterial diversity and space time dynamics in sugar beet on the north slope of Tianshan mountain, Appl. Microbiol. Biotechnol., 2014; 98: 6375-6385.
Crossref - Shylesh BS and Padikkala J. in vitro cytotoxic and antitumour property of E.sonchifolia (L.) in Mice, J.Ethnopharmacol., 2000; 73: 495-500.
Crossref - Strobel G and Daisy B. Bioprospecting for microbial endophytes and their natural products, Micobiol. Mol.Biol. Rev., 2003; 67: 491-502.
Crossref - Urumbil SK and Anilkumar M. Isolation, identification and characterization of plant growth promoting bacterial endophytes from E. sonchifolia (Linn.)DC., Asian J. Microbiol. Biotech. Env. Sc., 2019; 21(3): 121-128.
- vonMering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N and Bork P. Quantitative polygenetic assessment of microbial communities in diverse environments, Science, 2007; 315: 1126-1130.
Crossref - Yang R, Liu P and Ye W. Illumina based analysis of endophytic bacterial diversity of tree peony(Paeonia Sect. Mountain) roots and leaves, Brazilian Journal of Microbiology, 48(4): 695-705.
Crossref - Zam SI, Agustien A, Syamsuardi S, Djamaan A and Mustafa I. The diversity of endophytic bacteria from the traditional medicinal plants leaves that have anti-phytopathogens activity, Journal of Tropical Life Science, 2019; 9(1): 53-63.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.