ISSN: 0973-7510
E-ISSN: 2581-690X
Resistance against the routinely used antibiotics has reached a worrying level globally. Extended spectrum β-lactamases (ESBLs) production is the major mechanism of antimicrobial resistance. These ESBLs bacteria are resistance to penicillin, cephalosporins, monobactams. TEM1&2, CTX-M, SHV are the main ESBLs genes present in Klebsiella pneumoniae, which is produced by the alteration of amino acid in the active site. The aim of this study is to determine the prevalence of ESBL genes such as blaTEM 1&2, blaCTX-M and blaSHV. The present study was carried out from April 2019 to September 2019, a total of 121 K. pneumoniae isolates were collected and subjected to phenotypic study. Among these 19 isolated was ESBL positive, genes (blaSHV, blaTEM, blaCTX-M) were detected by conventional PCR method. blaTEM (100%) was the predominant gene detected flowed by CTX-M (68.42%) and SHV (57.89%). The highest level of antimicrobial resistance towards ampicillin (93.4%) followed by ceftriaxone (28.9%), cefotaxime (24.8%) and ciprofloxacin (22.3%). However, ESBL-producing isolates were showed resistance to ampicillin (100%) followed by ceftazidime (94.74%), cefotaxime (89.47%), amikacin and amoxicillin-clavulanic acid (68%). Antimicrobial resistance of bacteria is due to the genes, especially extended spectrum beta lactamase, which is widely found in members of Enterobacteriaceae. Nevertheless, there is a paucity of studies regarding the distribution of ESBL in K. pneumoniae in Palakkad Dist., Kerala. Hence the aim of the current study determines the distribution of ESBL genes in ESBL producing K. pneumoniae isolated from various clinical samples.
K. pneumoniae, Antimicrobial Resistance, ESBL, Genotype
Beta-lactam antibiotics are versatile antibiotics used for a broad spectrum of infections. The emergence of antimicrobial resistance is a significant threat in today’s society. The members of the Enterobacteriaceae family are commonly occurring bacteria in the environment. Klebsiella pneumoniae is one of the frequently isolating bacteria coming under this family. Resistance to various antibiotics is quite a concern because of the ESBL and carbapenem resistance of K. pneumoniae, which is repeatedly reported from different parts of the world. It is a causative agent of health care infections and community acquiring infections.1 The most common diseases are Urinary tract infections, respiratory tract infections, and wound infections, which now act as a co-pathogen in Covid -19 cases.
K. pneumoniae is a well-studied organism due to its multidrug resistance. The ESBL is an enzyme produced by bacteria, acting on the beta-lactam ring. Β-lactam antibiotics are widely used worldwide; they bind to the penicillin-binding protein and inhibit the biosynthesis of the bacterial cell wall. Nevertheless, these bacteria produce a β-lactamase enzyme, which acts on β- the lactam ring and develops resistance to beta-lactam antibiotics.2 The β -lactamase is classified by Ambler into four classes, A, B, C & D in this. Class A, C & D have serine enzymes in the active site and Zinc –Meltallo enzyme in the case of class B.3
ESBL gene is the essential resistance present in Enterobacteriaceae; it is encoded by blaTEM, blaCTX-M, and blaSHV genes, and of these, CTX-M is common. These plasmid-mediated genes are associated with other resistance genes, resulting in a co-resistance that includes aminoglycosides and tetracycline.4 The first ESBL resistance in K. pneumoniae was detected in Europe in 1980 and later spread to other areas. The distribution of ESBL resistance varies from one institution to another, one place to another. The ESBL resistance is a global concern; it leads to treatment failure, increased hospital stay, and a financial burden on humans. So the early detection of ESBL will help the clinicians with therapy. Hence, this study aims to detect the ESBL K. pneumoniae and the distribution of ESBL genes present in K. pneumoniae, isolated from different clinical samples.
The study was conducted from April 2019 to September 2019 in the Department of Microbiology, Karuna Medical College, Palakkad, Kerala. A total of 121 isolates of K. pneumoniae from different clinical samples were subjected to phenotypic studies.
The bacterial isolates were identified by biochemical reactions according to the standard protocol.5 All the K. pneumoniae isolates were phenotypically tested for ESBL production by double disc synergy test.6
Antimicrobial Susceptibility Testing
All ESBL-positive isolates were tested for antimicrobial susceptibility by the Kirby Bauer disc diffusion method according to CLSI guidelines.7 The discs used for susceptibility test, Ampicillin (10µg), cotrimoxazole (25µg), amoxyclave (10µg), cefotaxime (30µg), ceftriaxone (30µg), ceftazidime (30µg), ciproflaxacine (5 µg), amikacin (30µg), gentamicin (10µg), imipenem (10 µg) and tetracycline (30 µg), (Himedia, Mumbai).
ESBL Detection by Double Disc Synergy Test
The ESBL detection of all isolates is performed by double disc synergy test using cefotaxime (30 µl) and cefotaxime clavulanic acid (30 µl + 10 µl) combination according to CLSI guidelines.8 The K. pneumoniae isolates were tested with cefotaxime and ceftazidime disc with and without clavulanic acid are placed 25 mm apart from each other in Muller-Hinton agar (Himedia). The zone of inhibition >5mm in clavulanic acid combination is interpreted as ESBL positive.
Genotypic Method for the Detection of ESBL Gene
Phenotypically confirmed ESBL producing K. pneumoniae isolated were tested genotypically by conventional PCR. CTX-M, TEM 1&2, and SHV gene specific primers were used for performing PCR test. The plasmid DNA isolation was performed by using Thermo scientific TM Gene JET Plasmid Kit, according to manufactures instruction. The ESBL genes such as CTX-M Group 1, TEM 1 & 2, SHV were amplified individually by using specific primers (Table 1).
Table (1):
Primers used for PCR.
Gene |
Primers |
|---|---|
TEM 1 & 2 |
Forward Primer: 5′-CATTTYCGTGTCGCCCTTATTC-3′ Reverse Primer: 5′- CGTTCATCCATAGTTGCCTGAC-3′ |
SHV |
Forward Primer: 5′- ATCCCGCAGATAAATCACCAC-3′ Reverse Primer: 5′- AGCCGCTTGAGCAAATTAAAC-3′ |
CTX-M Group 1 |
Forward Primer: 5′- TTAGGAARTGTGCCGCTGYA-3′ Reverse Primer: 5′- CGATATCGTTGGTGGTRCCCAT-3′ |
Table (2):
Protocol for TEM 1 & TEM 2 Amplification.
| Step | Temperature | Time |
|---|---|---|
| Initial denaturation | 950C | 5 minutes |
| Denaturation | 940C | 30 seconds |
| Annealing | 620C | 40 seconds |
| Extension | 720C | 1 minute |
| Go to step 2-4 for 35 times | ||
| Elongation | 720C | 10 minutes |
Table (3):
Protocol for SHV gene amplification.
| Step | Temperature | Time |
|---|---|---|
| Initial denaturation | 950C | 5 minutes |
| Denaturation | 940C | 30 seconds |
| Annealing | 550C | 45seconds |
| Extension | 720C | 1 minute |
| Go to step 2-4 for 35 times | ||
| Elongation | 720C | 10 minutes |
Table (4):
Protocol for CTX-M gene Amplification.
| Step | Temperature | Time |
|---|---|---|
| Initial denaturation | 950C | 5 minutes |
| Denaturation | 940C | 30 seconds |
| Annealing | 580C | 45 seconds |
| Extension | 720C | 1 minute |
| Go to step 2-4 for 35 times | ||
| Elongation | 720C | 10 minutes |
The amplification mixture final volume 15µl containing, Thermo master mix 8 µl, primer mix 1µl, deionized water 5 µl and DNA template 1 µl. The PCR amplification condition of each gene displayed in Table 2,3 & 4. After amplification the PCR product were loaded in 1.5% agarose gel. After the run the band visualized on gel documentation system (Bio Rad gel doc XR+) and took the photographs.
Statistical Analysis
All the data were analyzed using SPSS version 21(IBM corporation/ Armonk, Newyork/ USA). The chi-square test and Fisher’s exact test evaluated the antimicrobial resistance patterns of ESBL and non-ESBL. ‘P’ value less than or equal to 0.01 was considered statistically significant.
In the present study, 121 K. pneumoniae were isolated from different clinical samples; among them, 58(47.9%) were females, and 63 (52.1%) were males (Table 5).
Table (5):
Gender wise distribution of K. pneumoniae isolates.
Gender |
Frequency |
Percent |
|---|---|---|
Female |
58 |
47.9 |
Male |
63 |
52.1 |
Total |
121 |
100.0 |
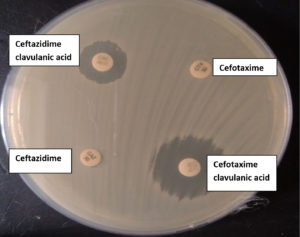
The distribution of isolates in different specimen, 56 (46.3%) urine, 37 (30.6%) sputum, 23(19%) pus, 2 (1.7%) blood and 1(0.8%) each from secretion tip, tissue and ascetic fluid (Table 6). Out of these 121 isolates, 19 were phenotypically positive for ESBL production (Figure 1). The distribution of ESBL positive isolates distribution among various clinical samples is shown in table 6, and among the clinical samples, ESBL prevalent in Urine 8 (14.3%), followed by sputum 5(13.51%) and pus 4 (17.4%).
Table (6):
Distribution of K. pneumoniae among the clinical sample.
No. |
Samples |
Number of isolates(N=121) |
Percentage (%) |
No; of ESBL positive isolates |
|---|---|---|---|---|
1 |
Urine |
56 |
46.3 |
8 (14.3) |
2 |
Sputum |
37 |
30.6 |
5 (13.51) |
3 |
Pus |
23 |
19.0 |
4(17.4) |
4 |
Blood |
2 |
1.7 |
0 (0) |
5 |
Secretion tip |
1 |
0.8 |
1 (100) |
6 |
Tissue |
1 |
0.8 |
1 (100) |
7 |
Ascitic fluid |
1 |
0.8 |
0 (0) |
8 |
Total |
121 |
(100) |
19 |
Table (7):
Comparison of antibiotic resistance patterns of ESBL and Non ESBL producing K. pneumoniae.
| Antibiotic | ESBL | c2 | P | ||
|---|---|---|---|---|---|
| Negative (n=102) |
Positive (n=19) |
||||
| AMP | Resistant | 94 | 19 | 1.596ns | 0.354ns(Fisher’s Exact test) |
| Susceptible | 8 | 0 | |||
| COT | Resistant | 8 | 9 | 20.72** | <0.001 |
| Susceptible | 94 | 10 | |||
| CTX | Resistant | 13 | 17 | 46.54** | <0.001 (Yates correction) |
| Susceptible | 99 | 2 | |||
| CAZ | Resistant | 8 | 18 | 66.63** | <0.001 (Yates correction) |
| Susceptible | 94 | 1 | |||
| CTR | Resistant | 19 | 16 | 33.39** | <0.001 |
| Susceptible | 83 | 3 | |||
| AK | Resistant | 8 | 13 | 40.98** | <0.001 |
| Susceptible | 94 | 6 | |||
| AMC | Resistant | 12 | 13 | 31.36** | <0.001 |
| Susceptible | 90 | 6 | |||
| GEN | Resistant | 10 | 10 | 21.29** | <0.001 |
| Susceptible | 92 | 9 | |||
| TE | Resistant | 8 | 12 | 35.52** | <0.001 |
| Susceptible | 94 | 7 | |||
| IMP | Resistant | 0 | 0 | – | – |
| Susceptible | 102 | 19 | |||
| CIP | Resistant | 16 | 11 | 16.46** | <0.001 |
| Susceptible | 86 | 8 | |||
| CX | Resistant | 17 | 10 | 11.951** | 0.002 |
** Significant at 0.01 level; ns Non significant
AMP-Ampicillin, COT- cotrimoxazole, CTX- cefotaxime, CAZ- ceftazidime, CTR- ceftriaxone, AK- amikacin, AMC- amoxicillin-clavulanic acid, GEN- gentamicin, TE- tetracycline, IPM- imipenem, CIP – ciprofloxacin, CX- cefoxitin
Figure 1. ESBL detection by double disc synergy test (CTX- Cefotaxime and CEC -Cefotaxime clavulanic acid; CAZ-Ceftazidime and CAC-Ceftazidime clavulanic acid)
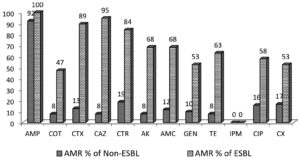
The antimicrobial resistance of ESBL and non-ESBL are shown in Figure 2. The percentage of resistance to cephalosporins in ESBL producing K. pneumoniae and non-ESBL producing K. pneumoniae were 89% and 13% for cefotaxime, 95% and 8% for ceftazidime, 84% and 19% for ceftriaxone. However, the ESBL and non-ESBLs were susceptible to carbapenem such as imipenem (100%).
Figure 2. Antimicrobial resistance in ESBL and Non-ESBL producing K. pneumoniae
AMP-Ampicillin, COT- cotrimoxazole, CTX- cefotaxime, CAZ- ceftazidime, CTR- ceftriaxone, AK- amikacin, AMC- amoxicillin-clavulanic acid, GEN- gentamicin, TE- tetracycline, IPM- imipenem, CIP – ciprofloxacin, CX- cefoxitin
The comparison of the antimicrobial resistance pattern of ESBL and non-ESBL is shown in Table 7. The association between antimicrobial resistance and ESBL production was statistically significant for Cotrimoxazole (<0.001), cefotaxime (<0.001), Ceftazidime (<0.001), ceftriazone (<0.001), amikacin (<0.001), amoxiclav (<0.001), gentamicin (<0.001), tetracycline (<0.001), ciprofloxacin (<0.001), cefoxitin (0.002). The antimicrobial susceptibility of ESBL isolates revealed that the highest resistance was against ampicillin (100%), followed by ceftazidime (94.74%) and cefotaxime (89.47%). All isolates were susceptible to imipenem.
** Significant at 0.01 level; ns Non significant
AMP-Ampicillin, COT- cotrimoxazole, CTX- cefotaxime, CAZ- ceftazidime, CTR- ceftriaxone, AK- amikacin, AMC- amoxicillin-clavulanic acid, GEN- gentamicin, TE- tetracycline, IPM- imipenem, CIP – ciprofloxacin, CX- cefoxitin
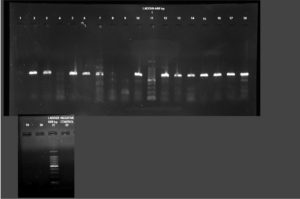
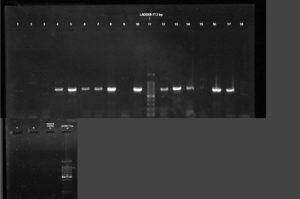
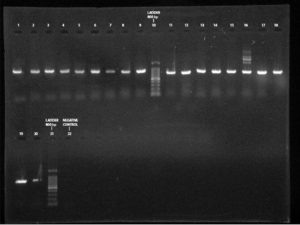
All phenotypically ESBL positive isolates were genotypically tested for the detection of resistance genes such as blaSHV, blaTEM 1 & 2, and blaCTX-M. Out of this beta-lactamase (bla) genes, all 19 (100%) isolates showed blaTEM 1 & 2, followed by blaCTX-M 13 (68.42%) and blaSHV 11(57.89%)(Figure 3-5). Among the nineteen K. pneumoniae isolates, 4 (21.05%) isolates possess only blaTEM 1 & 2 ESBL gene. No isolates carry blaCTX-M and blaSHV genes alone (Table 8).
Table (8):
Overview of ESBL genotype among K. pneumoniae isolates.
A. Single ESBL gene |
Number of positive |
Percentage (%) |
|---|---|---|
bla CTX-M |
0 |
0 |
bla TEM 1&2 |
4 |
21.05% |
bla SHV |
0 |
0 |
B. Two ESBL genes |
||
bla CTX-M and bla SHV |
0 |
0 |
bla CTX-M and bla TEM |
4 |
21.05% |
bla TEM and bla SHV |
2 |
10.53% |
C. Three ESBL genes |
||
bla CTX-M, bla TEM and bla SHV |
9 |
47.37 |
Figure 3. PCR amplification image of CTX-M gene in K. pneumoniae after 1.5% agarose gel electrophoresis
Figure 4. PCR amplification image of SHV gene in K. pneumoniae after 1.5% agarose gel electrophoresis
Antimicrobial resistance is a significant concern in the health care system. ESBL production is considered one of the antimicrobial resistance mechanisms in the family Enterobacteriaceae.9 ESBLs are the plasmid-coded genes, which provide resistance to third-generation cephalosporins, and monobactam (aztreonam)10 is considered an essential factor for the emergence of antimicrobial resistance.
Antimicrobial resistance in K.pneumoniae was noted as an important problem in the health care setting worldwide. In the present study, all isolates showed low susceptibility to ampicillin (6.61%) and a high-level susceptibility to imipenem (100%). This suggests ampicillin is inadequate for empirical treatment unless combined with other suitable drugs. In the current study, ESBL K. pneumoniae are resistant to amikacin (68%), gentamicin (53%), cotrimoxazole (47%), and ciprofloxacin (58%). The leading cause of antimicrobial resistance is the overuse of antibiotics, which is one of the factors contributing to the emergence of resistant bacteria in the community.11 Carbapenem is a high-level antibiotic used for the treatment of resistant organisms. The current study showed that all isolates are susceptible to imipenem. This follows Ghafourian et al.12 and Uddin et al.13
The distribution of ESBL-producing isolates varied between the geographical regions. In the present study, 19 (15.45%) ESBL K. pneumoniae were isolated phenotypically. This report follows the study from Hyderabad, India, conducted by Kumar et al.,14 showed 19% of ESBL in K. pneumoniae. A study from India reported that 25-84% was ESBL occurrence in different institutional studies.15 Another study from North India reported 52.27% of ESBL Klebsiella pneumoniae isolated from various clinical samples. A Study from China reported that ESBL producers’ isolation rate varies between 25-40%.16 Another study from Israel and Spain showed 40% ESBL prevalence in K. pneumoniae.17-18 Some other studies recorded a high incidence rate of ESBL-producing isolates, Feizabedi et al.,19 reported 69.7% of K. pneumoniae were ESBL positive from Iran, and a study by Lal et al.,20 noticed 97.1% of ESBL producers from India and another study from north India indicated 66.7% of K. pneumoniae were ESBL positive.21 Turkey (60%), Brazil (45.4%), Western Pacific (24.6%), Netherlands (22.6%), and Iran (44 -74%) have reported the highest rate of ESBL prevalence.22 The percentage of isolation of ESBL producers varies with different factors such as prolonged hospitalization, antibiotic use and policy, the hygiene of hospital personnel, and disinfection of ICU.23 Some studies reported a low level of ESBL prevalence in K. pneumoniae, 17%24-25 and 12%.26
The ESBL production is due to the mutation in TEM-1 & 2 and SHV 1 genes.27 In the current study, blaTEM (100%) was the most prevalent gene in ESBL producers, followed by CTX-M (68.42%) and SHV (57.89%). A similar result was found in the study conducted by Ferreiral et al.28 in this blaTEM (100%), CTX-M (72%), and SHV (96%). The ESBL genes were present in all ESBL positive isolates, but the prevalence of this gene varies. The previous reports from north India showed that 73% of blaTEM and 55% of blaCTX-M were the most dominant ESBL genes present in PGIMER and AIIMS, respectively. Another study from Lucknow also reported 75% of blaTEM in ESBL K. pneumoniae.29 Samanje et al.,30, in their study, found that 95% of ESBL K. pneumoniae isolates harbor the blaTEM gene, and 90% CTX-M were detected. On the other hand, over the past decade, CTX-M was the predominant gene reported in many Asian countries in K. pneumoniae.31 A study from Malaysia reported that the most prevalent ESBL gene in K. pneumoniae isolates was CTX-M (91.3%).32 A study from Nepal reported that 78.9% of K. pneumoniae contains the CTX-M gene.33 However, a study from the middle east detected that 93.75% (30/32) TEM and 87.5% (28/32) SHV gene was found in ESBL K. pneumoniae.34 The prevalence of the ESBL gene depends on various factors like the time of the study, the number of isolates, and geographical area.
The current study showed the presence of multiple genes in a single isolate. This follows the other studies.35 Nevertheless, the combination of ESBL genes is different in the different studies. The current study reported that the co-existence of blaTEM and blaCTX-M was 21.05% (4/19), and blaTEM and blaSHV was 10.53% (2/19). A study conducted by Bora et al.36 observed that 27.58% of K. pneumoniae isolates pose blaTEM and blaCTX-M together. In the present study, the presence of three genes was 47.37%. National Institute of hygiene in Lome, Togo, conducted a study from 2013-2015 that reported the presence of 3 genes in K. pneumoniae 59.26% (32/54).37 However, another study conducted by Manoharan et al.38 observed that 42.6% of K. pneumoniae isolates have all three genes TEM, SHV, and CTX-M. The co-existence of these three genes and some other antimicrobial resistance mechanisms may increase the antimicrobial resistance towards the last choice antibiotics like carbapenems and contribute to a high level of antimicrobial resistance.
ESBL is the most crucial antimicrobial resistant mechanism, and the high level of high resistance is consistent with the presence of the resistance gene in this study. Determining ESBL-producing bacterial isolates is essential in controlling infection and epidemiological surveillance studies. The current study reported that most ESBL-producing isolates are resistant to cephalosporins. The ESBL genes TEM 1 & 2, CTX-M, and SHV, these three genes are present in most cases. Only imipenem was active against the ESBL-producing K. pneumoniae. The knowledge, identification, and updating of the antimicrobial resistance among the common pathogens are helpful for the appropriate treatment of infections. Monitoring ESBL production using phenotypic and genotypic methods is recommended to benefit infection control.
ACKNOWLEDGMENTS
The authors would like to thank Principal, Karuna Medical College, Palakkad and Head, Department of Microbiology, Karuna Medical College for the facilities provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed in this study have made a substantial direct and intellectual contribution to the work and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated during the study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Nirwatil H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in tertiary care hospital, Indonesia. BMC Proc. 2019;13(Suppl 11):20.
Crossref - Umadevi S, Kandhakumari G, Joseph NM, et al. Prevalence and antimicrobial susceptibility pattern of ESBL producing Gram negative bacilli. J Clin Diagn Res. 2011;5(2):236-239.
- Bush K, Jacoby GA. Updated functional classification of β-lactamase. Antimicrob Agents Chemother. 2010;54(3):969-976.
Crossref - Tacao M, Moura A, Correia A, Henriques I. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic system. Water Res. 2014;48:100-107.
Crossref - Collee JG, Fraser AG, Marmian BP, Simmons A, Mackie and McCartney practical medical microbiology. 14th Ed. New York.Churchill Livingstone, 1996.
- Wayne PA. CLSI performance standard for antimicrobial susceptibility testing, Twenty-fifth informational supplement. Clinical and Laboratory Standards institute. CLSI document M100-S25; 2015.
- Wayne PA. CLSI performance standard for antimicrobial susceptibility testing. Twenty- fourth informational supplement. Clinical and Laboratory standards institute. CLSI document MI00-S24. 2014.
- Wayne PA. CLSI performance standard for antimicrobial susceptibility testing. Clinical and Laboratory standards institute. 30th ed. CLSI supplement MI00. 2020
- Thomson KS. Extended-spectrum-β- lactamase, AmpC and carbapenemase issue. J Clin Microbiol. 2010;48(4):1019-1025.
Crossref - Pitout JDD. Enterobacteriaceae that produce Extended- spectrum β- lactamases and AmpC β- lactamases in the community: the tip of the Iceberg?. Curr Pharm Des. 2013;19:257-263.
Crossref - Prestinanci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309-318.
Crossref - Ghafourian S, Sekawi Z, Neela V, Khosravi A, Rhabar M, Sadeghifard N. Incidence of Extended- spectrum beta- lactamases- producing Klebsiella pneumoniae in patients with urinary tract infection. Sao Paulo Med J. 2012;130(1):37-43.
Crossref - Uddin MA, Motazzim-Haque HM, Noor R. Isolation and identification of pathogenic Escherichia coli, Klebsiella spp. and Staphylococcus spp. in raw milk samples collected from different areas of Dhaka city, Bangladesh. Stamford Journal of Microbiology. 2011;1(1):19-23.
Crossref - Kumar MS, Lakshmi V, Rajagopalan R. Occurrence of Extended – spectrum beta- lactamases among Enterobacteriaceae spp isolated at a tertiary care institute. Indian J Med Microbiol. 2006;24(3):208-211.
Crossref - Kaur M. Occurrence of the CTX- M, SHV and the TEM genes among the extended spectrum beta lactamase producing isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. Journal of Clinical and Diagnostic Research. 2013: 642- 645.
Crossref - Yu Y, Zhou W, Chen Y, Ding Y, Ma Y. Epidemiological and antibiotic-resistant study on extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Zhejiang province. Chin Med J (Engl). 2002;115(10):1479-1482. PMID: 12490091
- Navon- Venezia S, Hammer-Munz O, Schwartz D, Turner D, Kuzmenko B, Carmeli Y. Occurrence and phenotypic characteristics of extended-spectrum beta- lactamase among members of the family Enterobacteriaceae at the Tel-Aviv Medical Center (Israel) and evaluation of diagnostic tests. J Clin Microbiol. 2003;41(1):155-158.
Crossref - Canton R, Novias A, Valverde A, et al. Prevalence and spread of extended – spectrum beta- lactamase- producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(1):144-53.
Crossref - Feizabadi MM, Delfani S, Raji N, et al. Distribution of bla (TEM), bla (SHV) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran. Iran Microb Drug Resist. 2010;16(1):49-53.
Crossref - Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM and SHV gene in extended – spectrum beta-lactamases (ESBLs) producing Klebsiella spp. Isolated from a tertiary care hospital. Indian J Med Res. 2007;125(2):173-178. PMID: 17431288.
- Goyal A, Prasad KN, Prasad A, Gupta S, Ghoshal U, Ayyagari A. Extended spectrum beta – lactamases in Escherichia coli and Klebsiella pneumoniae and associated risk factors. Indian J Med Res. 2009; 129(6): 695- 700. PMID: 19692752.
- Ghafourian S, Bin Sekawi Z, Sadeghifard N, et al. The prevalence of ESBLs producing Klebsiella pneumoniae isolates in some major hospitals, Iran. Open Microbiol J. 2011;5:91-95.
Crossref - Ananthan S, Subha A. Cefoxitin resistance mediated by loss of porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian J Med Microbiol. 2005;23(1):20-23.
Crossref - Tola MA, Abera NA, Gebeyehu YM, Dinku SF, Tullu KD. High prevalence of extended- spectrum beta-lactamases- producing Escherichia coli and Klebsiella pneumoniae fecal carriage among children under five years in Addis Ababa, Ethiopia. PloS ONE. 2021;16(10):e0258117.
Crossref - Riaz S, Faisal M, Hasnain S. Prevalence and comparison of beta-lactamase producing Escherichia coli and Klebsiella spp from clinical and environmental source in Lahore, Pakistan. Afr J Microbiolo Res. 2012;6(2):465-470.
Crossref - Behrooozi A, Rhabar M, Yousefi JV. Frequency of extended -spectrum beta- lactamase (ESBLs) producing Escherichia coli and Klebsiella pneumoniae isolated from urine in an Iranian 1000 -bed tertiary care hospital. Afr J Microbiol Res. 2010;4:881-884. http://www.academicjournals.org/ajmr
- Chaudhary U, Aggarwal R. Extended- spectrum beta-lactamases(ESBL)- an emerging threat to clinical therapeutics. Indian J Med Microbiol. 2004;22(2):75-80.
Crossref - Ferreial RL, da Silva BCM, Rezende GS, et al. High prevalence of multidrug- resistant Klebsiella pneumoniae harbouring several virulence and β- lactamase encoding genes in a Brazilian Intensive Care unit. Front Microbiol. 2019;9:3189.
Crossref - Jain A, Mondal R. TEM & amp; SHV genes in Extended-spectrum β- lactamase producing Klebsiella pneumoniae species beta their antimicrobial resistance pattern. Indian J Med Res. 2008;128(6):759-64. PMID: 19246801
- Samanje J, Mohammed AS, Al-Hamami MSS. Phenotypic and genotypic detection of Extended -spectrum β-lactamase production by Klebsiella pneumoniae isolated from different clinical sample in Baghdad, Iraq. J Pure Appl Microbiol. 2021;15(3):1681-1688.
Crossref - Shakya P, Shrestha D, Maharjan E, Sharma VK, Paudyal R. ESBL production among E. coli and Klebsiella spp. causing urinary tract infection: A hospital based study. Open Microbiol J. 2017;11(1):23-30.
Crossref - Al-Marzooq F, Yusof MYM, Tay ST. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLOS ONE. 2015;10(7):133654.
Crossref - Parajuli NP, Maharjan P, Joshi G, Khanal PR. Emerging perils of extended spectrum β -lactamase producing Enterobacteriaceae clinical isolates in a teaching hospital of Nepal. BioMed Res Int. 2016;2016:1782835.
Crossref - Aljanaby AAJ, Alhasani AHA. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr J Microbiol Res. 2016;10(22):829-843.
Crossref - Hassan H, Abdalhamid B. Molecular characterization of Extended- spectrum β- lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Ctries. 2014;8:282-288.
Crossref - Bora A, Hazarika NK, Shukla SK, Prasad KN, Sarma JB, Ahmed G. Prevalence of blaTEM , blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian J Pathol Microbiol. 2014;57(2):249-254.
Crossref - Diagbouga S, Salah FD, Sadji AY, et al. Detection of High Prevalence of TEM/SHV/CTX-M Genes in ESBL Producing and Multidrug Resistant Klebsiella pneumoniae and Klebsiella Oxytoca. J Clin Diagn Res. 2016;4(1):130.
Crossref - Manoharan A, Premalatha K, Chatterjee S, Mathai D. Correlation of TEM, SHV and CTX-M extended spectrum beta-lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011;29(2):161-164.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.