ISSN: 0973-7510
E-ISSN: 2581-690X
Ten sediment samples were gathered from several geographical locations around mangrove habitat, Red Sea coast, Egypt, during summer 2019. Actinobacteria are widespread in most mangrove soil samples. The average actinomycetes counts in sediment samples were ranged from 4 to 15 CFUg-1, also physico-chemical characters for soil samples were determined. Statistical analysis was applied to assess if the geographical location and physico-chemical characters influenced the communities of actinomycetes. A total of 10 actinomycetes were isolated and characterized physiologically and biochemically. The antimicrobial activities of different actinomycetes isolates were assessed. Isolate M3 was chosen as the most promising isolate with broad antagonistic activity against Bacillus subtilis ATCC 6633, Escherichia coli ATCC 19404, Staphylococcus aureus ATCC6538, Pseudomonas aeruginosa ATCC 9027, and Candida albicans ATCC 10231 with inhibition zones ranged from 12.0 ± 0.9 to 20.0 ± 1.9 mm. Genotypic characterization of isolate M3 was made using 16S rDNA sequence analysis and identified as Streptomyces mutabilis M3 with accession number MT483919. This strain exhibited anticancer activity against breast cancer cell line (Mcf7), liver cancer cell line (HepG2) and colon cancer cell line (HCT116) and the IC50 values were 324.77, 333.71 and 354.46, respectively. Streptomyces mutabilis M3 MT483919 had high bio-flocculating activity for seawater treatment, and the recovery of the samples ranged between 71.97 and 76.05%. The crude extract of Streptomyces mutabilis MT483919 M3 was analyzed by Fourier transform infrared spectrum (FT-IR) and Gas chromatography-mass spectrometry (GC-MS).
Mangrove Habitats, Streptomyces mutabilis, M3
Mangrove is a fertile and underexploited ecosystem with unlimited diversity of microbes for the detection of new and chemically diverse antimicrobial compounds1. Mangrove swamps are found in the world’s tropical areas which are sheltered from wave action and are usually highly sedimented. The forest of mangroves is a fringing population of shallow sandy or muddy areas, extending from the highest mark to the intertidal and subtidal regions2. Mangrove ecosystems are often inadequately explored but provide good resources to isolate novel actinomycetes3. Mangroves occur along the Red Sea coast and Sinai Peninsula in Egypt. At Gonah, about 25 km north of Hurghada, is the northernmost locality of mangroves along the Red Sea continental coast. From Hurghada to the Sudanese frontier, mangroves are prominent in protected bays, lagoons, beaches, dry wadis mouth and lacunae filled with sediments4. It is understood that climate fluctuations such as changes in salinity and tidal gradient are continuously encountered in the mangrove habitats. Forests of mangrove are home to numerous animals and plants5. Rather, these unexpected changes in the environment could be the motive force for the establishment of diversity of microbes and the variation in metabolic pathways that could create certain inimitable microbial properties6. Therore, the study of mangrove actinomycetes can offer a better view of discovering unique isolates, which lead to the discovery of respected antimicrobial compounds7,8. Scientistis are still studying the community diversity in phylum actinomycetes from diverse nations and environments, due to their ecological significance and biotechnological benefits9. Recent studies show that marine actinomycetes isolated from Red Sea mangrove forests can be a appreciated source for antibacterial, antioxidants and anticancer agents. The target of this study was to evaluate biodiversity and elucidate mangrove actinomycetes bioactivity along the mangrove area 17 km south of the Egyptian Red Sea coast town of Safaga city, and investigate the characterization of Streptomyces mutabilis M3 isolated from mangrove habitats and their antimicrobial and bio-loculation activities.
Gathering of Mangrove Soil Samples
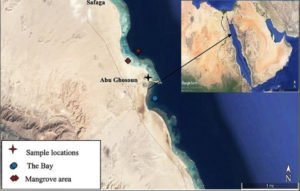
Ten soil samples were gathered from the selected sites in mangrove location during summer 2019. The mangrove area situated 17 km south of Safaga city on the western side of the Red Sea coast at the intersection of latitude 26°36΄53˝–26°37΄07˝N and longitudes 34°00΄46˝–34°00΄27˝E (Fig. 1). The samples locations and information are itemized in Table 1. The collected samples have been filled into sterilized packets and transported to the laboratory as soon as possible. All the mangrove forest areas studied are characterized by a single community dominated by Avicennia marina10.
Table (1):
Locations and Information of soil samples.
Sample number |
Location |
The characteristic of soil |
Sampling depth under surface (cm) |
|---|---|---|---|
1 |
26°36΄53˝ N-34°00΄49˝ E |
Rhizosphere soil of Avicennia marina |
5 |
2 |
26°36΄54˝ N-34°00΄46˝ E |
Rhizosphere soil of Avicennia marina |
5 |
3 |
26°36΄55˝ N-34°00΄45˝ E |
Rhizosphere soil of Avicennia marina |
10 |
4 |
26°36΄56˝ N-34°00΄43˝ E |
Muddy soil |
5 |
5 |
26°36΄57˝ N-34°00΄42˝ E |
Muddy soil |
10 |
6 |
26°36΄58˝ N-34°00΄40˝ E |
Sandy soil |
10 |
7 |
26°37΄01˝ N-34°00΄35˝ E |
Sandy soil |
10 |
8 |
26°37΄04˝ N-34°00΄30˝ E |
Rhizosphere soil of Avicennia marina |
10 |
9 |
26°37΄05˝ N-34°00΄30˝ E |
Muddy soil |
10 |
10 |
26°37΄06˝ N-34°00΄30˝ E |
Muddy soil |
10 |
Physico-chemical characteristics for samples
Physical parameters such as temperature, pH and salinity were examined for the soil samples using the 556 Handheld Multiparameter Instrument. Chemical characteristics such as dissolved oxygen12, dissolved nitrate13, dissolved nitrite, dissolved phosphate and dissolved ammonia14 were also measured using standard methods.
Isolation and purification of actinomycetes
The initial treatment of mangrove soil samples was based on the 60°C dry wet method in a combination of seawater (1: 1 v/v) for 15 minutes to reduce the number of unicellular bacteria 15. Soil suspension up to 1 ml was inoculated in Starch Casein Agar (SCA) medium (g/l: Starch, 10; Casein, 0.3; KNO3, 2; K2HPO4, 2; MgSO4.7H2O, 0.05; FeSO4.7H2O, 0.01; CaCO3, 0.02; Agar, 20) supplemented by nalidixic acid (20 mg/l), cycloheximide (50 mg/l), and potassium dichromate (50 mg/l) to prevent the growth of Gram-negative bacteria and fungi 1. After incubation at 32°C for 7-30 days. Colony Forming Unit (CFU) was estimated, colonies were selected up and streaked on SCA medium. Pure cultures have been maintained on SCA agar slants at 4°C for numerous weeks and have also been well-maintained in suspensions (20 %, v / v) of glycerol at −80°C.
Characterization of actinomycetes isolates
Out of 65 actinomycetes isolates, 10 isolates were selected based on their color variability and pigmentation on starch casein agar medium and incubation 7-14 days at 32°C. Morphology of colonies of the isolates was detected regarding color of substrate and aerial mycelia in addition to branching according to16. Cultural characteristics (growth, aerial and substratum mycelium coloration, soluble pigment formation) have been tested in seven different media including Yeast extract malt extract agar17, Inorganic salts starch agar 18, Czapex Dox agar19, Krassilnikov agar20, Oat meal agar21 and Starch nitrate agar22.
Biochemical characterization
Biochemical tests generally used are hydrolysis of starch, casein, urea and gelatin. In addition to indole production test, methyl red test, Voges-prauskauer test, nitrate reduction, H2S production, catalase, oxidase and gelatin liquefaction tests23
Physiological characterization
Physiological characterization, such as optimum pH (5-9), temperature (15-50°C) and salinity (NaCl 0-13%) were also tested. Use of carbon sources such as sucrose, fructose, glucose, starch, dextrose and maltose. Also some sources of nitrogen such as L-tyrosine and L-arginine were tested24.
Fermentation
The chosen actinomycetes isolates were then cultured on soluble Starch Casein media with pH of (7.5). The selected actinomycetes were inoculated in the prepared medium and retained at room temperature for 4–7 days in a rotary shaker. The cultures were then centrifuged for 15 min at 8000 rpm, and the supernatant was obtained for extra studies25.
Antimicrobial activity
The bacterial pathogens used in the present study were Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC 6633, Pseudomonas aeruginosa ATCC 9027, Salmonella typhimurium ATCC 14028, Escherichia coli ATCC 19404 and Candida albicans ATCC 10231. The pathogenic strains were kindly gained from The National Institute of Oceanography and Fisheries (NIOF), Egypt. The antibacterial activities of the actinomycetes’ supernatant was evaluated using well cut diffusion technique. The pathogens were inoculated by using sterilized cotton swabs on Mueller Hinton agar. Next, wells were bored using a well-borer sterilized micro tip and 100 µl of crude extracts were added into the wells. The plates were incubated overnight at 37°C. Duplicate was prepared for each set. After incubation, positive result was taken as the radius in (mm) of clear inhibition zone around each well26.
Solvent extraction
The bioactive metabolites (s) in the crude extract was extracted using ethyl acetate. The extraction was done by mixing 1:1 ratio of the crude extract and the solvent in separate funnel and was vigorously shaken for 1h then left to stand for 24 h. Further extraction of the organic layer was done for additional studies27.
Molecular identification of actinomycetes
Genomic DNA of representative selected actinomycetes isolate were extracted using Gene Jet genomic DNA purification Kit (Fermenats) using the manufacturer’s protocol. The 16S rRNA gene PCR amplification was performed using the following primers: 27F: (5’ AGAGTTTGATCCTGGCTCTCAG–3’) and 1492R (5’GGTTACCTTGTTACGACTT–3’)28. The final volume of PCR reactions was 50 μl consisting of 200 ng DNA templates, 25 μl Maxima Hot Start PCR Master Mix (2Xs) and 0,4 μM primers under the following conditions: initial 94° denaturation for 5 minutes followed by 30 s at 94°C cycles for 30s, 60s at 55°C and 4 minutes extension phase at 72°C and 10 minutes extension point. PCR reactions were conducted in a 50 μl final volume. The amplification products were confirmed using 1% agarose gel and sent to GATC Biotech Company, German for purification and sequencing. The GenBank BLASTn search tool (http://www.ncbi.mlm.nih.gov) was used to analyze the partial nucleotide selected isolate sequences. This supported the isolate closest phylogenetic neighbors. The GenBank submitted the following accessionMT483919number to partial 16S rRNA gene sequence of actinomycetes isolated. The Sequence was then aligned by the software Clustal W, and the phylogenetic tree by molecular Evolutionary genetic analysis (MEGA version 6.0) was extracted from a neighboring joining algorithm.
Anticancer activity
The cytotoxic effect of Streptomyces mutabilis M3. extract on breast cancer cell line (Mcf7), liver cancer cell line (HepG2) and colon cancer cell line (HCT116)- was investigated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Cells were grown in 96-well plate at density of 1x 105cell/ml and incubated for 24 h at 37 °C to form monolayer sheets. The growth medium was decanted and the cells were washed twice and 0.1 mL of RPMI containing different concentration (31.25 to 1000 ϻg/ ml) of the extract was inoculated in each well. Control wells were treated with the medium containing the solvent (DMSO) without sample. After incubation at 37 °C with 5% CO2 for 72 h, 20 ϻL of MTT solution (5mg/ml in PBS) was added to each well, mixed thoroughly and further incubated for 1-5 h. the medium was removed and Formazan was dissolved in 100 ϻl of DMSO. The amount of MTT-formazan was determined at 570 nm absorbance with 650 nm as reference wavelength29-31.
Bio-flocculation activity
A protocol of Kurane32 is used to study the bio-flocculating activity, Briefly, 2 ml of the culture broth, 5 ml of CaCl2(1%, w/v), and 93 ml of kaolin (5 gl-1 distilled water) suspension were added in a 200- ml beaker and stirred at 180 r min−1 for 1.5 min and at 80 r min−1 for 3 min with a vortex mixer (QL- 861, Shanghai Jingmi Instrument Co., Ltd., China) and then kept still for 10 min. A sample was withdrawn (1cm below the surface of clay suspension) to measure optical densities (OD). A control experiment without bio-flocculant was carried prepared. The bio-flocculation activity was determined relying on OD for the suspension of clay using spectrophotometer at 550 nm. The flocculating efficiency was calculated using the following equation: Bio-flocculation activity = (B-A)/B×100. Where; A and B are ODs of the culture sample and the control, respectively.
Fourier transform infrared (FT-IR) spectral analysis
Functional groups of the extracted compound was determined using FT-IR spectrometer in the scan range of 450 to 4000 cm−1. AV500 FT-NMR spectrometer was used for H1NMR analysis to recognize the functional groups exist in the compound33.
Gas chromatography-mass spectrometry (GC-MS) analysis
The GC-MS (Jeol GCMATE II GC-MS) analysis was carried out and the device is attached to a component of the mass spectrum (MS) system and Agilent Technologies 6890 NGC system for gas chromatography.HP5 (50 m to 0.25 mm) of GC column was used. For a period of 20 minutes the temperature of the column was maintained at 100°C, 3 min was maintained at 235°C and the injector temperature was 240°C. The results of the MS analysis match the library and identify the closest match34.
Statistical analysis
Significant analysis, canonical correspondence analysis (CCA) was determined using XLSTAT, 2014 to find the association between three sets of variables in 1000 permutations and a 5% significance level. Actinomycetes count was the first variable, distribution of sites was the second variable and physico-chemical parameters were the third variable. Principle coordinate analysis (PCO) was determined by XLSTAT, 2014 for distributing the correlated sites according to actinomycetes communities and physico-chemical parameters35.
Distribution of marine actinomycetes in mangrove soil samples
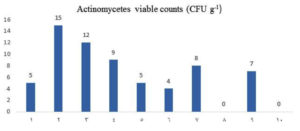
The average actinomycetes counts (CFU g-1) were estimated during the summer of 2019 in soil samples from mangrove area, Red Sea coast, Egypt (Fig. 2). In general, the sample number 2 showed higher counts of actinomycetes (15 CFU g-1), whereas the lowest counts (4 CFU g-1) reported in sample number 6. Actinobacteria are widespread in all mangrove environments1,36,37. Several previous studies reports the occurrences of actinomycetes in different mangrove habitats from various geographical locations around the world38. Previous study showed 85 actinobacterial isolates from 60 sediment samples collected from mangrove rhizosphere on the Egyptian Red Sea coast39.
Physicochemical characters of soil samples
The variations in environmental factors (temperature, pH, salinity, dissolved oxygen, dissolved nitrate, dissolved nitrite, dissolved phosphate and dissolved ammonia) of soil samples lead to variations in marine actinomycetes density (Table 2). Isik40 and his team, reported that, actinomycetes present in any sample would be significantly influenced by geographical location and environmental factors.
Table (2):
Physicochemical characters of soil samples collected from mangrove area during Summer 2019.
| Sample number |
Physical-chemical parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| Temperature (°C) |
pH | Salinity (‰) |
DO (mgl-1) | NO3 (µM) |
NO2 (µM) |
NH4 (µM) | PO4 (µM) |
|
| 1 | 32.1 | 8.75 | 42.1 | 6.3 | 0.5 | 0.02 | 3.06 | 0.05 |
| 2 | 32.2 | 8.71 | 42.1 | 6.5 | 0.3 | 0.08 | 2.73 | 0.04 |
| 3 | 32.3 | 8.81 | 42.0 | 6.5 | 0.3 | 0.07 | 2.84 | 0.04 |
| 4 | 32.1 | 8.69 | 42.2 | 6.8 | 0.4 | 0.07 | 2.87 | 0.05 |
| 5 | 32.4 | 8.77 | 42.0 | 6.5 | 0.3 | 0.08 | 2.88 | 0.04 |
| 6 | 32.4 | 8.78 | 42.2 | 6.6 | 0.5 | 0.09 | 2.67 | 0.08 |
| 7 | 32.5 | 8.72 | 42.1 | 6.4 | 0.4 | 0.04 | 2.94 | 0.04 |
| 8 | 32.3 | 8.78 | 42.0 | 6.5 | 0.9 | 0.08 | 3.09 | 0.08 |
| 9 | 32.1 | 8.69 | 42.1 | 6.5 | 0.5 | 0.06 | 2.99 | 0.04 |
| 10 | 32.2 | 8.73 | 42.2 | 6.1 | 0.7 | 0.09 | 3.13 | 0.05 |
Canonical correspondence analysis and principle coordinate analysis
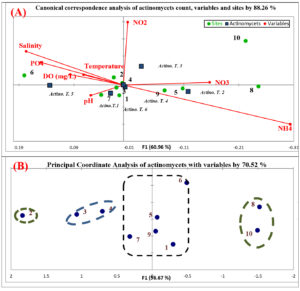
Influence of physico-chemical parameters on actinomycetes community was assessed using Canonical correspondence analysis (CCA). The community of actinomycetes achieved 88.26% similarity with physico-chemical parameters and stations origin diversity. Based on Fig. (3-A), actinomycetes communities influenced by pH, temperature, nitrate, nitrite and dissolved oxygen. At the station level, samples 1, 3, 4 and 7 are positively stimulated by pH. Samples 5, 8 and 9 were positively stimulated by nitrate. Samples 2 and 6 were positively stimulated by temperature, salinity, phosphate and dissolved oxygen. Actinomycetes are act as neutrophils in marine ecosystem, so pH is very important factor for enumeration, distribution and activity of these microorganisms41. In the present study, samples were alkaline in all the stations and this leading to increase the actinomycetes count. The dominant pH for growing actinomycetes is neutral pH 742. This is different in these studies which all actinomycetes pH ranged from 8-9 pH. Beside pH, temperature is second most important factor for microbial growth. Actinomycetes are mesophilic and growing in temperature range (25 – 30°C). The range of temperature was relatively higher in the current study and the influence of temperature on distribution of the actinomycetes in the present study was observed. Similar results were reported by Abd-Elnaby et al.43. Nitrate and nitrite of different stations is different, and this influences the communities of actinomycetes. NO2 is relatively unsteady and can be quickly oxidized to NO3. Higher concentrations of NO2 can arise in the distribution system when chlorination is used, but the occurrence is always irregular. The concentration of NO3 and NO2 in different sites is relatively in acceptable range and they affected positively on growth of actinomycetes communities44. The increasing of water dissolved oxygen is indicator for microbial decomposition of organic matters. So, the recording of actinomycetes in samples is influenced by amount of organic matter45. This is clearly summarized by dissolved oxygen range in study area. The second statistical analysis (principle coordinate analysis) illustrated the correlation between samples and different parameters with actinomycetes count (Fig. 3-B). Samples 1, 5, 6, 7 and 9 were correlated by 20%. The nature of soil in these sites is different, but they are similar in influencing by different variables. On the other hand, samples 3, 4, 8 and 10 were correlated by 40%. Finally, sample 2 is not correlated with any other sites upon correlation of different parameters. This refers to the origin of parameters in this site.
Fig. 3. Statistical analysis of actinomycetes, sites and physico-chemical parameters; (A)Biplot of canonical correspondence analysis (ccA); (B) Biplot of principle coordinate analysis (PCoA)
Morphological, biochemical and physiological characterization
Of the total marine actinomycetes isolates obtained, ten actinobacterial isolates were selected and coded as (M1, M2, M3, M4, M5, M6, M7, M8, M9 and M10). They were selected on the basis of leathery powdery growth, color variability and pigmentation. Table 3 reported the cultures and characteristics of marine actinomycetes isolates. Aerial mass color of substrate mycelium was determined after 7 to 10 days. All marine actinomycetes isolates grown well on all tested media except three isolates (M6, M8 and M9) could not grow on starch agar and inorganic salts. The substrate mycelium of five isolates was yellow, three isolates had cream color and two isolates had Pink. Most of them had white or gray aerial mycelium. Two isolates produce beige diffusible pigments, only one produce violet and other one produce dark brown. All isolates grew at 25-30°C and the majority grew at 35°C, pH 7-9. Starch, lactose, fructose, dextrose, glucose and sucrose were utilized by all strains as carbon sources, the majority utilized maltose. Most of them utilized l-arginine and l-tyrosine. All isolates produced catalase and the majority produced gelatinase. Indeed, characterization studies help to explain biological, ecological, physiological and biochemical characteristics. The characterization is as important as an analysis of the existence of these microbes 46. This is of utmost importance for understanding the basic physiology of the marine isolates of actinomycete to be biochemically and physiologically characterized47.
Table (3):
Phenotypic characteristics of actinomycetes isolates.
| Character | Actinomycetes isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | ||
| Growth on | |||||||||||
| Yeast extract malt extract agar | + | + | + | + | + | + | + | + | + | + | |
| Inorganic saltsstarch agar | + | + | + | + | + | – | + | – | – | + | |
| Czapex Dox agar | + | + | + | + | + | + | + | + | + | + | |
| Krassilnikov agar | + | + | + | + | + | + | + | + | + | + | |
| Oat meal agar | + | + | + | + | + | + | + | + | + | + | |
| Starch nitrate agar | + | + | + | + | + | + | + | + | + | + | |
| Starch casein agar | + | + | + | + | + | + | + | + | + | + | |
| Substrate mycelium | |||||||||||
| Yellow | – | – | + | + | – | – | – | + | + | + | |
| Cream | + | – | – | – | – | + | + | – | – | – | |
| Pink | – | + | – | – | + | – | – | – | – | – | |
| Aerial mycelium | |||||||||||
| White | + | + | – | – | – | – | – | + | + | + | |
| Cream | – | – | – | – | – | – | – | – | – | – | |
| Gray | – | – | + | + | – | + | – | – | – | – | |
| Yellow | – | – | – | – | – | – | + | – | – | – | |
| Pink | – | – | – | – | + | – | – | – | – | – | |
| Brown | – | – | – | – | – | – | – | – | – | – | |
| Diffusible pigments | |||||||||||
| Beige | – | – | + | – | – | – | + | – | – | – | |
| Yellow | – | – | – | – | – | – | – | – | – | – | |
| Violet | – | + | – | – | – | – | – | – | – | – | |
| Dark brown | – | – | – | + | – | – | – | – | – | – | |
| Growth at (°C) | |||||||||||
| 15-25 | + | + | + | + | + | + | + | + | + | + | |
| 30-35 | + | + | – | + | + | + | + | + | – | + | |
| 40-50 | – | – | – | – | – | – | – | – | – | – | |
| Growth at pH | |||||||||||
| 5-6 | – | – | – | – | + | – | – | – | – | – | |
| 7-9 | + | + | + | + | + | + | + | + | + | + | |
| Utilization of | |||||||||||
| Starch | + | + | + | + | + | + | + | + | + | + | |
| Lactose | + | + | + | + | + | + | + | + | + | + | |
| Fructose | + | + | + | + | + | + | + | + | + | + | |
| Dextrose | + | + | + | + | + | + | + | + | + | + | |
| Maltose | – | + | – | + | + | + | + | + | – | + | |
| Glucose | + | + | + | + | + | + | + | + | + | + | |
| Sucrose | + | + | + | + | + | + | + | + | + | + | |
| L-arginine | + | – | + | + | – | – | + | + | – | – | |
| L-tyrosine | + | – | + | + | + | – | + | + | + | – | |
| Growth in presence of NaCl (%) | |||||||||||
| 0-4 | – | – | – | – | – | + | – | – | + | – | |
| 7-10 | + | + | + | + | + | + | + | + | + | + | |
| 13 | – | – | – | – | – | – | – | – | – | – | |
| Biochemical testes | |||||||||||
| Casein hydrolysis | + | + | + | + | + | + | + | + | + | + | |
| Urea hydrolysis | + | – | + | – | – | – | – | + | – | – | |
| Gelatin hydrolysis | + | + | + | + | – | + | – | + | – | – | |
| Indole production | – | – | – | – | – | – | – | – | + | – | |
| Methyl red | – | – | – | – | – | – | – | – | – | – | |
| Voges prauskauer | – | – | – | – | – | – | – | – | – | – | |
| H2S production | – | – | – | – | – | – | – | – | – | – | |
| Catalase production | + | + | + | + | + | + | + | + | + | + | |
Antimicrobial activities
Assessment of the antimicrobial activity of the cell-free supernatant of the selected ten marine actinomycetes isolates using the agar well diffusion method against tested pathogens was demonstrated as in Table 4. Significantly, the actinomycetes isolate M3 showed the highest inhibition zones 12.0 ± 0.9, 20.0 ± 0.7, 18.0 ± 0.9, 20.0 ± 1.9 and 12.0 ± 1.2 mm against Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC 6633, Pseudomonas aeruginosa ATCC 9027, Escherichia coli ATCC 19404 and Candida albicans ATCC 10231, respectively (Fig. 4). Actinomycetes isolate M5 showed 14.0 ± 1.6, 12.0 ± 0.5, 16.0 ± 1.2 and 20.0 ± 0.8 mm against Staphylococcus aureus ATCC6538, Pseudomonas aeruginosa ATCC 9027, Salmonella typhimurium ATCC 14028 and Escherichia coli ATCC 19404, respectively. On the other hand, isolate M9 affected on three pathogens only, isolates M1, M2, M4, M6, and M8 affected on two microbial pathogens, finally isolates M7 and M10 affected on one pathogen.
Table (4):
Antimicrobial activity of actinomycetes isolates against different pathogens.
| Inhibition zone (mm) | ||||||
|---|---|---|---|---|---|---|
| Isolates | S. aureus | B. subtilis | P. aeruginosa | S. typhimurium | E. coli | C. albicans |
| M1 | 10.0 ± 1.2 | 0.0 | 0.0 | 0.0 | 8.0 ± 1.6 | 0.0 |
| M2 | 16.0 ± 1.4 | 0.0 | 0.0 | 0.0 | 16.0 ± 1.6 | 0.0 |
| M3 | 12.0 ± 0.9 | 20.0 ± 0.7 | 18.0 ± 0.9 | 0.0 | 20.0 ± 1.9 | 12.0 ± 1.2 |
| M4 | 0.0 | 12.0 ± 0.8 | 0.0 | 16.0 ± 1.2 | 0.0 | 0.0 |
| M5 | 14.0 ± 1.6 | 0.0 | 12.0 ± 0.5 | 16.0 ± 1.2 | 20.0 ± 0.8 | 0.0 |
| M6 | 0.0 | 16.0 ± 1.7 | 0.0 | 0.0 | 12.0 ± 1.9 | 0.0 |
| M7 | 10.0 ± 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M8 | 0.0 | 0.0 | 10.0 ± 0.6 | 0.0 | 12.0 ± 2.1 | 0.0 |
| M9 | 14.0 ± 1.4 | 0.0 | 12.0 ± 1.3 | 0.0 | 20.0 ± 0.5 | 0.0 |
| M10 | 0.0 | 16.0 ± 2.1 | 0.0 | 0.0 | 0.0 | 0.0 |
Mangrove microorganisms, in particular actinobacteria, were reported to be capable of producing structurally unique bioactive natural products1,36. Previous study reported the presence of bioactive compounds (particularly antibiotics) in actinomycetes inhabiting the mangrove environment38,48. For examples, crude extracts of actinomycetes isolates (A5, A71 and A70) exhibited strong antimicrobial activity39 Streptomyces sp. isolate EGY2 showed highest antimicrobial and cytotoxic activity against MDA-MB-231 breast cancer cell line49.
Molecular phylogeny of the selected isolate
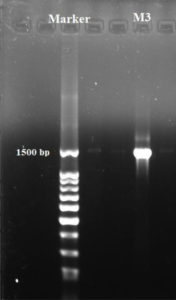
Based on the results obtained, isolate M3 has been selected for molecular phylogenetic analysis and identification using16S rDNA comparison sequence phylogenetic analysis in order to create phylogenetic and evolutionary organismal relations50. Agarose gel electrophoresis (Fig. 5) has been used to detect the produced amplicons from the isolate selected.
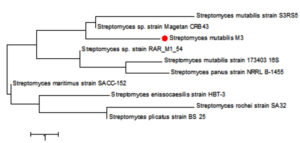
The sequencing data utilizing this strategy (ABI 3730xl) was 1500 base pair, and it was aligned with other sequences of related actinomycetes on the database to determine its phylogenetic relationship to other actinomycetes. According to 16S rRNA gene sequence analysis of isolate M3 compared to Blast search computer-based program which provided the highest homology. The results showed that, the isolate under study was similar to Streptomyces mutabilis with 97% similarity percentage. The sequence of isolate M3 was deposited in Gene-Bank as Streptomyces mutabilis M3 with accession number MT483919, and the phylogenetic tree was constructed as shown in (Fig. 6). The identification of Streptomyces sp. using 16S rRNA revealed that the marine soil samples are predominantly insulated. This result is in line with previous research, where Streptomyces sp. recovery was higher than the other actinobacterial genera with antimicrobial activity 51. Streptomyces mutabilis sp. MII. which was previously isolated from a sediment sample collected in the Red Sea at the Hurghada Coast has new bioactive compounds and showed good antimicrobial and anticancer activity52.
Fig. 6. Phylogenetic tree based on 16S rRNA sequences using neighbor joining method showing relationships between the selected M3 isolate isolated from mangrove habitat and their closely related type strains
Anticancer activity
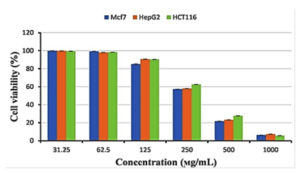
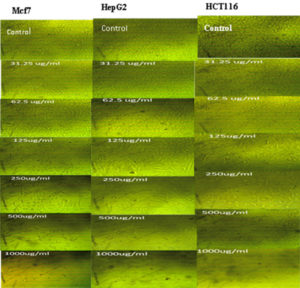
The cytotoxicity of Streptomyces mutabilis M3 MT483919 extract was evaluated toward different human cancer cell lines using MTT assay. Fig. 7 shows the cell viability of the different cancer cell lines after exposure to various concentrations of the extract for 72 h. The extract exhibited a significant reduction in the viability of Mcf7, HepG2 and HCT116 cancer cell lines and the IC50 (concentration that necessary to cause 50% reduction in cell viability were 324.77, 333.31 and 354.46, respectively. In addition, Streptomyces mutabilis M3 extract altered the morphology of the viable cancer cell lines tested (Fig. 8). Actinomycetes are rich source of unmatched anticancer compounds specially the Streptomyces because they possess different gene clusters responsible for the manufacture of polyketide and no ribosomal peptide synthases53,54. Many studies have confirmed the antitumor activity of different Streptomyces species55. The extract of Streptomyces mutabilis M3 reduced the viability of Mfc7, HepG2 and HCT116 cancer cell lines by more than 90% at concentration 1000 ϻg/ml. These results suggest that the strain produce secondary metabolites with anticancer activity and the purified components of the extract could be more promising in terms of activity and the effective concentration.
Fig. 7. Cytotoxicity of Streptomyces mutabilis M3 MT483919 extract against breast cancer cell line (Mcf7), liver cancer cell line (HepG2) and colon cancer cell line (HCT116)
Fig. 8. Morphology of Mfc7, HepG2 and HCt116 cancer cell lines after exposure to different concentrations (0-1000 ϻg/mL) of Streptomyces mutabilis M3 MT483919 extract for 72 h
Bio-flocculating activity
Treatment of wastewater is a main research topic worldwide. Flocculation is regarded as amazing way for eliminating pollutants from wastewater as was previously reported56. Primarily Streptomyces mutabilis extract showed considerable bio- flocculating activity in kaolin clay treatment (Fig. 9) so that, it had remarkable efficiency when tested as a bio-flocculant producer for treatment of different seawater samples collected from different sites (Ebrahemia, Eastern Harbor and Quit-Bay) along Mediterranean Sea shores, Alexandria, Egypt. The results in Table 5 explained that, Streptomyces mutabilis M3 MT483919 has high bio flocculating activity and the recovery of the samples ranged between 71.97 and 76.05 Similar results were reported by57 who reported the promising applications of the bio flocculant QZ-7 synthesized by a novel Bacillus salmalaya strain 139SI in wastewater treatment.
 Fig. 9. Bio-flocculant activity by Streptomyces mutabilis M3 MT483919 extract. The sample after treatment (A) and before treatment (control) (B)
Fig. 9. Bio-flocculant activity by Streptomyces mutabilis M3 MT483919 extract. The sample after treatment (A) and before treatment (control) (B)
Table (5):Bio-flocculant activity of Streptomyces mutabilis M3 MT483919 extract for seawater treatment.
| Sea water sample | OD at 550 nm | Bio-floculating activity % | |
|---|---|---|---|
| Before treatment (Control) | After treatment | ||
| Ebrahemia | 1.645 | 0.394 | 76.05 |
| Eastern Harbor | 1.795 | 0.434 | 75.82 |
| Quit-Bay | 1.905 | 0.534 | 71.97 |
Chemical Composition analysis of crude extract
Fourier transform infrared (FT-IR) spectral analysis
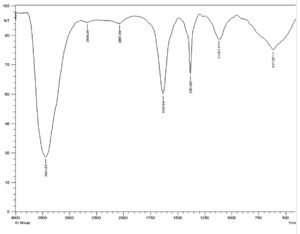
The FT-IR spectrum of antimicrobial substance produced by isolate M3 showed peaks at 3441.01 cm−1 ,2669.48 cm−1, 2067.69 cm−1, 1635.64 cm−1, 1381.03 cm−1, 1118.71 cm−1 and 617.22 cm−1 ,corresponding to the following functional groups such as alcohol (O-H), alkanes (C-H), alkene (C=C), amine(N-H ), alcohol (O-H), amine (C-N) and halo compound (C-Br), respectively (Fig. 10).
Similarly, salim and his team reported absorbance ranging from 3359.18 to 2927.10 cm−1 for the isolate of actinomycetes FA9, corresponding to amine group58. A peak of 3398 cm-1 characteristic of hydroxyl (O-H) and 3 common vibration peaks between 2899 and 2977 cm-1 characteristic of (C-H) symmetric (saturated) vibration of hydrocarbon was also recorded in previous study59.
GC-MS analysis
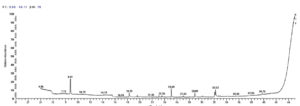
The GC-MS study has been used to classify the bioactive components. A total of 16 peaks are shown in the chromatogram GC-MS of the extract (Fig. 11). The GC-MS crude extract chromatogram identified 17 peaks at various times and retention times. They were found with the use of the NIST database (NIST 11 MS Server and MS Search System v.2.0 g).The active principles with their retention time (RT), molecular formula (MF), Molecular weight (MW) and concentration (%) are accessible in Table 6. The major compounds were Undecane, 2H-Pyran-3-ol,tetrahydro-2,2,6–trimethyl–6-(4-methyl-3-cyclohexen-1-yl)-,[3S-[3alpha,-6alpha (R)]]and 9-Octadecenoic acid (Z)- methyl ester with Peak area percentage 30.68, 11.48 and 10.86 %, respectively.
Fig. 11. GC-MS chromatogram and structure formulas of compounds of the bioactive compounds extracted from Streptomyces mutabilis M3 MT483919
Table (6): Components detected in Streptomyces mutabilis M3 MT483919 crude extract.
No |
R T Value (In Min.) |
Compound |
Mol. Formula |
Molecular Weight |
Peak area (%) |
|---|---|---|---|---|---|
1 |
5.74 |
Diisopropylamine |
C6H15N |
101 |
3.10 |
2 |
8.81 |
Undecane |
C11H24 |
156 |
30.68 |
3 |
14.19 |
Ethanol, 2-(diethylamino) |
C6H15NO |
117 |
1.92 |
4 |
15.50 |
Acetone, 1-[4- (dimethylaminoethoxy)phenyl]- |
C13H19NO2 |
221 |
3.98 |
5 |
18.30 |
1,2-Benzenedicarboxylic acid, dimethyl ester |
C10H10O4 |
194 |
7.42 |
6 |
19.58 |
Benzoic acid, 3-methyl-2-trimethylsilyloxy-, trimethylsilyl |
C14H24O3S2 |
296 |
2.73 |
7 |
23.08 |
Bisabolol oxide B |
C15H26O2 |
238 |
2.23 |
8 |
23.56 |
9,12-octadecadienoic acid |
C27H54O4Si2 |
498 |
2.48 |
9 |
23.71 |
Z,Z,Z-1,4,6,9-Nonadecatetraene |
C19H32 |
260 |
1.26 |
10 |
25.09 |
2H-Pyran-3-ol, tetrahydro-2,2,6-trimethyl-6-(4-methyl-3-cyclohexen-1-yl)-, [3S-[3alpha,6alpha(R)]]- |
C15H26O2 |
238 |
11.48 |
11 |
27.02 |
9,12,15-octadecatrienoic a |
C27H52O4Si2 |
496 |
2.06 |
12 |
28.89 |
Hexadecanoic acid, methyl |
C17H34O2 |
270 |
7.52 |
13 |
32.10 |
11,14-Octadecadienoic acid, methyl ester |
C19H34O2 |
294 |
6.70 |
14 |
32.22 |
9-Octadecenoic acid (Z)-, methyl ester |
C19H36O2 |
296 |
10.86 |
15 |
32.71 |
Cyclopropanebutanoic acid |
C25H42O2 |
374 |
1.46 |
16 |
39.75 |
1-Monolinoleoylglycerol trimethylsilyl ether |
C27H54O4Si2 |
498 |
2.67 |
Many of the compounds found have shown interesting biological activities60. Antibacterial activity is reported to be caused by hexadecanoic acid, octadecanic acid and tetradecanoic acid61. Octadecanoic acid collected from neem extract was examined against Salmonella sp., S. aureus and E. coli) and showed good inhibition activity against S. aureus than E. coli and Salmonella spp.62. Selvin and his team observed hexadecanoic acid and hexadecanoic acid methyl ester from actinomycetes isolate Nocardiopsisda ssonvillei MAD08 by using GC–MS analysis63. The basic components of GC-MS from Cytosoriacompressa are the fatty acids (hexadecanoic and octadecanoic acid) and fatty acid esters (hexadecanoic hydroxy ester and hexadecanic acids)64. A novel Streptomyces isolate Chy 2-3 isolated from chyulu National Park produced bioactive metabolites was identified as Octadecanoic acid that has antifungal, antitumor and antibacterial activity65.
In conclusion, reasonable numbers of actinomycetes isolates were successfully isolated from mangrove collected from 17 km south of the Egyptian Red Sea coast town of Safaga. Some of the isolates investigated showed wide range of antimicrobial activity against the tested indicator pathogenic bacteria. Future research including study of valuable biotechnological applications of the extracted active metabolites will be conducted.
ACKNOWLEDGMENTS
We are grateful to the management of the NIOF Red Sea Branch and Field Strategy Trips for the studying of mangrove environment and biodiversity project.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the National institute of oceanography and fishers, Egypt, fund via 2019 Research Plans Support Program.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the author.
AVAILABILITY OF DATA
The data used to support the findings of this study are available from the corresponding authors upon request.
- Li F, Liu S, Lu Q, et al. Studies on antibacterial activity and diversity of cultivable actinobacteria isolated from mangrove soil in Futian and Maoweihai of China. Based Complement and Alternat Med. 2019;2019.
Crossref - Dawes C. Marine Botany. JohnWiley&Sons. New York, NY, USA. 1981;160.
- Rajkumar J, Swarnakumar N, Sivakumar K, Thangaradjou, T, Kannan L. Actinobacterial diversity of mangrove environment of the Bhitherkanika mangroves, east coast of Orissa, India. International Journal of Scientific and Research Publications. 2012;2:1-6.
- Abd El-Wahab E. Autecology and Conservation Considerations of Avicennia marina (Forsk.) Vierh. in the Gulf of Aqaba, Egypt, M. Sc. Thesis, Faculty of Science, Zagazig University, Zagazig. 2004.
- Sathya R, Ushadevi T. Industrially important enzymes producing streptomyces species from mangrove sediments. Int J Pharm Pharm Sci. 2014;6:233-237.
- Law JWF, Ser HL, Khan TM, et al. Streptomyces colonosanans sp. nov., a novel actinobacterium isolated from Malaysia mangrove soil exhibiting antioxidative activity and cytotoxic potential against human colon cancer cell lines. Front Microbiol. 2017;8:877.
Crossref - Tiwari K, Gupta RK. Diversity and isolation of rare actinomycetes: an overview. Critical Rev Microbiol. 2013;39(3):256-294.
Crossref - Lee LH, Zainal N, Azman AS, et al. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int J Syst Evol Microbiol. 2014;64(Pt 9):3297-3306.
Crossref - Fu PC, Zhang YZ, Geng HM, Chen SL. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. Peer J. 2016;4:e2540.
Crossref - Madkour HA, Mohammed AW, . Nature and geochemistry of surface sediments of the mangrove environment along the Egyptian Red Sea coast. Environ Geol. 2008;54:257-267.
Crossref - Hasan MH. Destruction of sea cucumber populations due to overfishing at Abu Ghosoun area, Red Sea. J Basic Appl Zool. 2019;80:5.
Crossref - Rice EW, Baird RB, Eaton AD, Clesceri LS. Standard methods for the examination of water and wastewater. (American Public Health Association Washington, DC. USA. 2012;10.
- Strickland J, Parsons T. Bull. Fish. Res. Bd Can. A practical handbook of seawater analysis. 1968;311.
- Koroleff F, Grasshoff K. Methods of seawater analysis. GRASSHOFF, K. ed. 1976.
- Retnowati Y, Sembiring L, Moeljopawiro S, Djohan TS, Soetarto ES. Diversity of antibiotic-producing actinomycetes in mangrove forest of Torosiaje, Gorontalo, Indonesia. Biodiversitas J Biol Div. 2017;18:1453-1461.
- Bensultana A, Ouhdouch Y, Hassani L, Mezrioui NE, Rafouk L. Isolation and characterization of wastewater sand filter actinomycetes. World J Microbiol Biotechnol. 2010;26:481-487.
Crossref - Bawazir AMA, Shivanna GB, Shantaram M. Ipact of different media for growth and production of different pigments in actinomycetes isolated from soils of Hadhramout, YEMEN. Eur J Biomed. 2018;5:615-619.
- Maria A, Sharmili A, Anbumalarmathi J. Isolation and characterization of actinomycetes from marine soil. MOJ Biol Med. 2018;3:221-225.
- Marshall KC, Alexander M. Growth characteristics of fungi and actinomycetes. J Bacteriol. 1960;80(3):412-416.
Crossref - Krassilnikov N, Koreniako A. The bactericidal substance of the actinomycetes. Microbiol. 1939;8:673-685.
- Jiang Y, Li Q, Chen X, Jiang, C. Isolation and cultivation methods of Actinobacteria. Actinobacteria-basic Biotechnol Appll. 2016;39-57.
Crossref - Kumar RR, Jadeja VJ. Isolation of actinomycetes: A complete approach. Int J Curr Microbiol Appl Sci. 2016;5(5):606-618.
Crossref - Chaudhary HS, Yadav J, Shrivastava AR, Singh S, Singh AK, Gopalan N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India). J Advanced pharma Technol Res. 2013;4(2):118-123.
Crossref - Remya M, Vijayakumar R. Isolation and characterization of marine antagonistic actinomycetes from west coast of India. Medicine Biol. 2008;15:13-19.
- Priyanka S, Jayashree M, Shivani R, Anwesha S, Rao KB. Characterisation and identification of antibacterial compound from marine actinobacteria: In vitro and in silico analysis. J Infect Public Health. 2019;12(1):83-89.
Crossref - Malek NA, Zainuddin Z, Chowdhury AJK, Abidin ZAZ. Diversity and antimicrobial activity of mangrove soil actinomycetes isolated from Tanjung Lumpur, Kuantan. J Teknol. 2015;77(25):37-43.
Crossref - Jayasudha J, Kumar G, Karthik L, Bhaskara Rao K. Biological control of vibriosis by antagonistic actinobacteria-an in vitro study. J Agricul Technol. 2011;7:271-280 .
- Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clinic microbiol. 1990;28(9):1942-1946.
Crossref - Slater T, Sawyer B, Strauli U. Studies on succinate-tetrazolium reductase systems: III. Points of coupling of four different tetrazolium salts III. Points of coupling of four different tetrazolium salts. Biochim Biophysica Acta. 1963;77:383-393.
Crossref - Van de Loosdrecht A, Beelen R, Ossenkoppele G, Broekhoven M, Langenhuijsen MA. A Tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J immunol Methods. 1994;174(1-2):311-320.
Crossref - Alley MC, Scudiero DA, Monks A, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589-601.
- Kurane R, Hatamochi K, Kakuno T, Kiyohara M, Hirono M, Taniguchi Y. Production of a bioflocculant by Rhodococcus erythropolis S-1 grown on alcohols. Biosci Biotechnol Biochem. 1994;58(2):428-429.
Crossref - Ravi L, Kannabiran K. Extraction and Identification of gancidin W from marine Streptomyces sp. VITLGK012. Indian J Pharma Sci. 2018;80:1093-1099.
- Gnanavel V, Saral AM. GC-MS analysis of petroleum ether and ethanol leaf extracts from Arbus precatorius linn. Int J Pharm Bio Sci. 2013;4(3):37-44.
- Belal AAM, Kelany MS, Hamed MM, Abd El-Fattah LS. Selected bacterial communities associated with macro-benthic fauna assemblages at the Timsah Lake and the Western Lagoon’s sediments, Suez Canal, Egypt. Egypt J Aquatic Res. 2020;46(2):137-143.
Crossref - Xu DB, Ye WW, Han Y, Deng ZX, Hong K. Natural products from mangrove actinomycetes. Mar Drugs. 2014;12(5):2590-2613.
Crossref - Sun C, Li F, Cai Z, Wu L. Brief introduction of mangrove researches including drug discovery granted by National Natural Science Foundation of China from 1986 to 2016. Chin J Antibiot. 2017;2:241-248.
- Rosmine E, Varghese SA. Isolation of actinomycetes from mangrove and estuarine sediments of Cochin and screening for antimicrobial activity. J Coastal Life Med. 2016;4:207-210.
Crossref - Beltagy E, Abdel-Shakour S, Qari S H, Reffat B. Diversity and antimicrobial activity of Red Sea mangrove actinomycetes (Egypt). Int J Environ Sci Eng. 2015;6:59-74 .
- Isik K, Gencbay T, Ozdemir-Kocak F, Cil E. Molecular identification of different actinomycetes isolated from East Black Sea region plateau soil by 16S rDNA gene sequencing. Afr J Microbiol Res. 2014;8(9):878-887.
Crossref - Das S, Lyla P, Khan SA. Distribution and generic composition of culturable marine actinomycetes from the sediments of Indian continental slope of Bay of Bengal. Chin J Ocean Limnol. 2008;26:166-177.
Crossref - Selyanin V, Oborotov G, Zenova G, Zvyagintsev D. Alkaliphilic soil actinomycetes. Microbiol. 2005: 74:729-734.
Crossref - Abd-Elnaby HM, Abo-Elala GM, Abdel-Raouf UM, Hamed MM. Distribution and characterization of Actinomycetes in Suez Bay sediments. Egypt Asian J Appl Sci. 2016;4.
- Rabeh SA, Azab EA, Aly MM. Studies on bacterioplankton and inhibitory strains of aquatic actinomycetes in Lake Bardawil, Egypt. World J Microbiol Biotechnol. 2007;23:167-176.
Crossref - Walter SRS. Jaekel U, Osterholz, H, et al. Microbial decomposition of marine dissolved organic matter in cool oceanic crust. Nat Geosci. 2018;11:334-339.
Crossref - Hirsch AM, Valdes M. Micromonospora: An important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem. 2010;42:536-542.
Crossref - Deepthi A, Rosamma P. Actinomycete isolates from Arabian Sea and Bay of Bengal: biochemical, molecular and functional characterization, Cochin University of Science And Technology. 2014.
- Tan LTH, Chan KG, Pusparajah P, et al. Mangrove derived Streptomyces sp. MUM265 as a potential source of antioxidant and anticolon-cancer agents. BMC Microbiol. 2019;19(1):38.
Crossref - Abdelfattah MS, Elmallah MIY, Hawas UW, El-Kassema LTA, Eid MAG. Isolation and characterization of marine-derived actinomycetes with cytotoxic activity from the Red Sea coast. Asian Pacific J Tropical Biomed. 2016;6(8):651-657.
Crossref - Sharma M, Dangi P, Choudhary M. Actinomycetes: source, identification, and their applications. Int J Curr Microbiol App Sci. 2014;3:801-832.
- Dasn R, Romi W, Das R, Sharma HK, Thakur D. Antimicrobial potentiality of actinobacteria isolated from two microbiologically unexplored forest ecosystems of Northeast India. BMC Microbiol. 2018;18:71.
Crossref - Hamed A, Abdel-Razek AS, Frese M, et al. Acetylborrelidin B: a new bioactive metabolite from Streptomyces mutabilis sp. MII. Z Naturforsch C. 2018;73:49-57.
Crossref - Lopez-Lazaro MM, Pastor N, Azrak SS, Ayuso MJ, Austin CA, Cortes F. Digitoxin inhibits the growth of cancer cell lines at concentrations commonly found in cardiac patients. J Nat Prod. 2005;68(11):1642-1645.
Crossref - Aftab U, Zechel DL, Sajid I. Antitumor compounds from Streptomyces sp. KML-2, isolated from Khewra salt mines, Pakistan. Biol Res. 2015;48:58.
Crossref - Hussain A, Rather MA, Dar MS, et al. Streptomyces puniceus strain AS13., Production, characterization and evaluation of bioactive metabolites: A new face of dinactin as an antitumor antibiotic. Microbiol Res. 2018;207:196-202.
Crossref - Agunbiade MO, Pohl CH, Ashafa AO. A Review of the Application of Biofloccualnts in Wastewater Treatment. Polish J Environ Studies. 2016;25(4):1381-1389.
Crossref - Abu Tawila, ZM, Ismail S, Dadrasnia A, Usman MM. Production and characterization of a bioflocculant produced by Bacillus salmalaya 139SI-7 and its applications in wastewater treatment. Molecules. 2018;23(10):2689.
Crossref - Salim FM, Sharmili SA, Anbumalarmathi J, Umamaheswari K. Isolation, molecular characterization and identification of antibiotic producing actinomycetes from soil samples. J Appl Pharma Sci. 2017;7:069-075.
- Priya AJ, Sagadevan E, Arumugam. Detection of antioxidant and antimicrobial activities in marine actinomycetes isolated from Puducherry coastal region. J Modern Biotechnol. 2012;1:63-69.
- Belakhdar G, Benjouad A, Abdennebi E. Determination of some bioactive chemical constituents from Thesium humile Vahl. J Mater Environ Sci. 2015;6:2778-2783.
- Agoramoorthy G, Chandrasekaran M, Venkatesalu V, Hsu M. Antibacterial and antifungal activities of fatty acid methyl esters of the blind-your-eye mangrove from India. Brazilian J Microbiol. 2007;38(4):739-742.
Crossref - Pu ZH, Zhang Y-Q, Yin Z-Q, et al. Antibacterial activity of 9-octadecanoic acid-hexadecanoic acid-tetrahydrofuran-3, 4-diyl ester from neem oil. Agricul Sci China. 2010;9(8):1236-1240.
Crossref - Selvin J, Shanmughapriyan S, Gandhimathi R, et al. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes Nocardiopsis dassonvillei MAD08. Appl Microbiol Biotechnol. 2009;83(3):435-445.
Crossref - Abou-Elela GM, Abd-Elnaby H, Ibrahim HA, Okbah M. Marine natural products and their potential applications as anti-infective agents. World Appl Sci J. 2009;7:872-880.
- Karanja E, Boga H, Muigai A, Wamunyokoli F, Kinyua J, Nonoh J. “Growth characteristics and production of secondary metabolites from selected novel Streptomyces species isolated from selected Kenyan national parks,” in Proceedings of the 5th JKUAT Scientific, Technological and Industrialization Conference (Juja: Jomo Kenyatta University of Agriculture and Technology). 2010;51-80.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.