ISSN: 0973-7510
E-ISSN: 2581-690X
Food contains several microorganisms that may cause illnesses and food poisoning in humans. Small numbers of microorganism contamination could result in rapid spoilage of food. The Centres for Disease Control and Prevention (CDC), USA estimates that 76 million people are affected by foodborne illnesses each year in the USA. Salmonella infections alone account for one billion dollars yearly in direct and indirect medical costs and more than 5,000 deaths. In Sudan, diarrhoeal disease was reported as the second major disease during the years from 2003 to 2007 (Annual health statistical report of the Federal Ministry of Health, Sudan). We aimed to develop a rapid molecular procedure for the detection of Escherichia coli, Shigella dysentery, and salmonella Typhiin food so as to minimize the public health hazard of food contamination. We used the Multiplex PCR method as rapid methods were tested for identification of Enterobacteriaceae species Escherichia coli as an indicator organism for food contamination and two strains of Enterobacteriaceae that causes food borne illness (namely Shigella dysentery and salmonella Typhi). The Multiplex PCR was performed to detect E. coli using Mdh primer pair, Salmonella Typhi using IpaB primer pair, and Shigella dysentery using IpaH1 primer pair. The sensitivity to detect E. coli, Salmonella Typhi, and Shigella dysentery in contaminated food in the concentration of the infective and the over infective doses were 100%, 96.3%, and 88.9% respectively for the three bacteria strains. There was no significant difference in the detection of the bacteria after incubation for 8 hours, 24 hours, or even without incubation period. There were no differences in the result of the samples that were contaminated artificially in laboratory and those obtained from the market. The Multiplex PCR method for identification of E. coli, Salmonella Typhi and Shigella dysentery was developed as a model for detection and risk assessment of the three bacteria in one program, and it is suitable for routine analysis.
Bacterial Species, Food Inspection, Multiplex-PCR
Foodborne illness is one of the major global public health problems.1 Food gets contaminated via soil, water, sewage, and air as well as by contact with other plants and animals. Additional contamination of foods occurs during any time of production and preparation of food.2 Laboratory protocols are a vital tool in case investigation. Enterobacteriaceae is one of the important groups of foodborne pathogens.3 Enterobacteriaceae is Gram-negative facultative anaerobic bacillus, which ferment glucose to be acidic under anaerobic conditions, often with gas production. They are non-spore formers, short rods in shape, motile by peritrichous flagella or non-motile.4 Enterobacteriaceae may cause infectious diseases.5 The most important members of the Enterobacteriaceae family are Escherichia coli, Shigella, and Salmonella.
In Sudan, several studies isolated Enterobacteriaceae; E. coli among other species were isolated by Hussein6 from fresh meat samples; Abd ElRahman7 isolated E. coli from 17% of minced meat samples; Mohamed8 found that 6% of meat samples were E. coli beneficial; Ahmed8 and Ahmed9 identified E. coli in ready to eat beef burger in Khartoum State and found that the total coliform bacteria observed in 48.8% of samples and 9.3% of them were positive for E. coli. Warsama et al.11 isolated E. coli in 11.1% of food samples, and Salmonella Typhi in 11.1% of them. Moreover, Hassan11 identified E. coli among other species of bacteria from restaurant food, food handlers and food utensils in Khartoum State, Sudan. CDC Bulletin12 confirmed the presence of Salmonella in Australia and there was an epidemiological link with other travelers from other countries (including Sudan). Mustafa and Abdallah13 identified E. coli and Salmonella spp. in 6.6%, and 5% respectively from Street-Vended Um-Jingir (traditional food) in Khartoum State, Sudan. 14
Rapid Methods of Enterobacteriaceae Identification
The latest technology assists in detection and identification of microorganisms promptly with convenient use of the procedure, with high sensitivity rates and more accurate than the conventional techniques. Most of the methods used to detect specific pathogens in foods require some growth in an enrichment medium before analysis. Some rapid methods can be done in a few minutes to a few hours, so they yield results more quickly than traditional methods. However, in food analysis, rapid methods still lack sufficient sensitivity and specificity for direct testing; hence, foods still need to be culturally enriched before analysis. Although enrichment is a limitation in terms of technique speed, it provides essential benefits, such as diluting the effects of inhibitors, allowing the differentiation of viable from non-viable cells, and allowing for the repair of cell stress or injury that may have resulted during food processing.15
Several studies were achieved at a global level to improve the detection of food microorganisms aimed to ensure the quality and safety of food.16 Overviews of rapid methods for the detection of bacteria were done17,18; and Wang et al.19 These overviews include the progress and application of impedimetric biosensors for the detection of foodborne pathogenic bacteria, particularly the new specific bio-recognition elements such as bacteriophage, the use of nano-materials, and micro-fluidics techniques. Abubakar
et al.19 wrote a review on the public health and cost-effectiveness of rapid diagnostic tests for the detection and identification of bacterial intestinal pathogens in food.
Conventional polymerase chain reaction (PCR)
Polymerase Chain Reaction (PCR) is an in vitro method for amplification of targeted nucleic acid fragment (DNA) using a heating source (thermocycler), and a pair of primers, a template (thermostable DNA polymerase) and dNTPs.20 It has been used extensively since 1985 to rapidly detect, characterize, and identify a variety of organisms by detecting the presence of target gene fragments. PCR is a widely used tool in molecular biology to identify nucleic acid sequences.21,22
In this study, we aimed to develop a rapid method(multiplex PCR) for the detection of food contaminated with (E. Coli, Salmonella Typhi, and Shigella dysentery), so as to minimize the public health hazard of food contamination.
Bacterial strains
The bacterial strains used throughout this study were reference strains bacteria from the American Type Culture Collection (ATCC), Japan Collection of Microorganism (JCM), Korean Collection for Type Culture (KCTC), National Health Lab, Federal Ministry of Health, Sudan (NHL) as shown in Table (1).
Table (1):
Bacterial Strains used in this study.
No |
Bacterial strain |
Code |
|---|---|---|
1 |
Escherichia coli |
ATCC 25922 |
6 |
Salmonella Typhi |
ATCC 122235 |
10 |
Shigella dysentery |
NHL |
All bacterial cells were cultured and then lyophilized until they were used.
DNA extraction
DNA extraction was carried out according to the boiling cells method.
Primers
A total of six sets of primers were chosen: two sets of primers for each strain of three bacteria as mentioned in (Table 2).
Table (2):
Primer sequences of species.
| Bacterial species | Primer | Sequence(5’-3’) | Size (bp) | Reference |
|---|---|---|---|---|
| E. coli | Mdh |
F: ACTGAAAGCCAAACAGCCAAG | 392 | 24 |
| R:CGTTCTGTTCAAATGCGCTCAGG | ||||
| EC
|
F: ATCACCGTGGTGACGCATGTCGC | 486 | 25 | |
| R: CACCACGATGCCATGTTCATCTGC | ||||
| Salmonella spp* | IpaB | F: GGACTTTTTAAAAGCGGCGG R: GCCTCTCCCAGAGCCGTCTGG |
314 | 24 |
| invA | F: GTG AAA TTA TCG CCA CGT TCG GGC AA R: TCA TCG CAC CGT CAA AGG AAC C |
284 | 26, 27 | |
| Shigella spp* | IpaH
|
F: CCTTGACCGCCTTTCCGATAC R: CAGCCACCCTCTGAGAGTACTC |
600 | 24 |
| IpaH2
|
F: CGCAATACCTCCGGATTCC R: biotin-TCCGCAGAGGCACTGAGTT |
65 | 23 |
* spp= species
PCR protocol
Amplification of target DNA sequences was performed in a 20 μl reaction mixture in clean, sterile 0.2 ml polypropylene microcentrifuge tubes. The reaction mixture (per tube) consisted of 9.2 μl ddH2O, 2.5 μl 10x PCR Tris-acetate EDTABuffer33 (100 mm Tris-HCl, 500 mm KCl, 15 mm MgCl2, pH 8.3), 1.0 μl dNTP mixture, 1.0 μl forward primer, 1.0 μl reverse primer, 0.3 μl Taq polymerase (5U), and 5 μl DNA template as shown in Table (3). The reaction mixture was placed in a thermocycler under the following conditions: Initial temperature at 95°C for 5 minutes, followed by 35 cycles of: (denaturation at 94°C for 1 min, primer annealing at 59°C for 1min, and extension at 72°C for 1min), an additional step at 5 min at 72°C for primer extension was added at the end of the reaction, then hold at 4°C. DNA amplicons were analyzed by gel electrophoresis. The gel used in all experiments was 1.5%. Agarose electrophoresis was performed on Thermo EC gel trays. Gels were stained with ethidium bromide (Fisher Biotech, BP1302-10). DNA amplicons and DNA markers were visualized by a UV transilluminator. Images of DNA amplicon bands were captured using a photo documentation digital camera. 23,24
Table (3):
The samples of contaminated food.
| Food | Number of samples |
|---|---|
| Yogurt | 2 |
| Milk | 4 |
| Egg | 4 |
| Bread | 2 |
| Meat | 2 |
| Chicken | 2 |
| Fish | 2 |
| Beef burger | 2 |
| Sausage | 2 |
| Tomato
Carrot |
2 |
| 3 | |
| Total samples | 27 |
Identification of contaminated samples using PCR technique
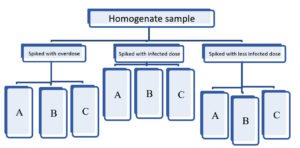
Twenty-seven food samples were purchased from the retail market at Khartoum Locality, Sudan, and contaminated by serial dilution of bacterial strains as shown in (Table 3).
25 grams of each sample were placed into a stomacher sterile bag with 225 ml buffer peptone water, and samples were blended at 230 rpm for 2 min using a Stomacher (Stomacher 400, Seward, and Norfolk, UK).
Strains and dilution
Three dilutions were done for each bacterial strain. The sample from the market was:
- Contaminated with over infectious dose of three bacteria.
- Contaminated with the infectious dose of three bacteria.
- Contaminated with less than the infectious dose of three bacteria.
Bacterial inocula used in spiking food portions (over-infectious, infectious dose and less than infectious dose as log10of colony-forming units as showing in Table (4).
Table (4):
Bacterial inocula used in spiking food portions, expressed as log10 of colony-forming units.
Bacteria |
Over infectious dose |
Infectious dose |
Less than the infectious dose |
Reference |
|---|---|---|---|---|
E. coli |
107 |
105 |
103 |
28 |
Salmonella Typhi |
107 |
105 |
103 |
28-30 |
Shigella dysentery |
103 |
10 |
<10 |
28,29 |
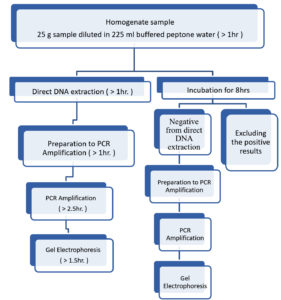
Fig. 1. DNA extraction from the contaminated food samples
A – Extract DNA directly without incubation period.
B- Extract DNA after incubated at 37°C for 8 hrs.
C- Extract DNA after incubated at 37°C for 24 hrs.
Isolation of DNA from contaminated samples
From each contaminated food sample there was three DNA Extraction. The DNA extraction from the contaminated food samples is shown in (Figure 1). These three DNA extractions were:
- from contaminated food sample directly without incubation period.
- from contaminated food sample after incubating at 37°C for 8 hours.
- from contaminated food sample after incubating at 37°C for 24 hrs.
So each contaminated sample has nine DNA extractions for PCR.
Bacterial DNA was detectable at different infection concentration levels (less infected, infected, and over infected). The three bacterial species E. coli, Salmonella Typhi, and Shigella dysentery were highly detectable at contaminated concentration levels tested as shown in Tables (5, 6 and 7).
Table (5):
PCR detection of E. coli in various artificially contaminated foods with different dilutions of E. coli.
| Type of food | Less than infectious doze | Infectious doze | Over infectious dose | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 2 | 100 |
| Milk | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Egg | 4 | 3 | 75 | 4 | 4 | 100 | 4 | 4 | 100 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Fish | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomato
Carrot |
2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| 3 | 2 | 66.7 | 3 | 3 | 100 | 3 | 3 | 100 | |
| Total | 27 | 24 | 88.9 | 27 | 27 | 100 | 27 | 27 | 100 |
E. coli was identified in all the contaminated samples tested except for yogurt and eggs in which the percentage of positive samples were 50% and 75% respectively as shown in Table (5).
Salmonella Typhi was identified efficiently, in high percentages even in concentrations of less than infectious dose. The detection level reaches 100% for all types of food tested except for tomatoes and carrots in which, Salmonella had been detected in 50% and 66.7 % of samples respectively. In yogurt, the detection level decreased to 50% of samples at the infectious dose concentration. However, yogurt showed a 100% detection level in less infectious and over the infectious concentrations. The overall detection level is 96.3% (Table 6).
Table (6):
PCR result of different kinds of food contaminated with a different dilution of Salmonella Typhi.
| Type of food | Less than infectious dose | Infectious doze | Over infectious dose | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 2 | 100 | 2 | 1 | 50 | 2 | 2 | 100 |
| Milk | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Egg | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Fish | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomatoes
Carrots |
2 | 1 | 50 | 2 | 2 | 100 | 2 | 2 | 100 |
| 3 | 2 | 66.7 | 3 | 3 | 100 | 3 | 2 | 66.7 | |
| Total | 27 | 25 | 92.6 | 27 | 26 | 96.3 | 27 | 26 | 96.3 |
Shigella dysentery was identified in less percentage at all dilutions compared to Salmonella Typhi except in bread, meat, chicken, and beef burger samples. At infectious and over infectious dozes the percentage was raised to 88.9% (Table 7).
Table (7):
PCR result of different kinds of food contaminated with a different dilution of Shigella dysentery.
| Type of food | Less than infectious dose | Infectious dose | Over infectious dose | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 1 | 50 |
| Milk | 4 | 3 | 75 | 4 | 4 | 100 | 4 | 4 | 100 |
| Egg | 4 | 3 | 75 | 4 | 3 | 75 | 4 | 3 | 75 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 1 | 50 | 2 | 2 | 100 |
| Fish | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 1 | 50 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomato
Carrot |
2 | 1 | 50 | 2 | 2 | 100 | 2 | 2 | 100 |
| 3 | 2 | 66.7 | 3 | 2 | 66.7 | 3 | 3 | 100 | |
| Total | 27 | 20 | 74.1 | 27 | 24 | 88.9 | 27 | 24 | 88.9 |
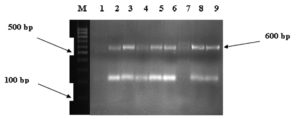
We obtained1.5% Agarose gel electrophoresis of PCR products from DNA extracted from pure bacteria. Lanes M, molecular size markers (100 bp ladder); lane 1 negative control; lane 7 negative result; and lanes 2,3,4,5,8, and 9 positive Shigella dysentery IpaH gene result (Fig. 2).
Different incubation periods
DNA from the three bacterial species was detectable by the multiplex PCR in all food samples after the different incubation periods. The results of contaminated food at different incubation periods are illustrated in Tables (8, 9 and 10). Bacterial DNA was detectable at three different incubation periods for all the food tested. Vegetables and yogurt showed a less degree of detection.
Table (8):
PCR result of E. coli for contaminated foods after different incubation periods.
| Type of food | Directly tested without incubation | Tested after 8 hrs Incubation |
Tests of overnight culture | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Milk | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Egg | 4 | 4 | 10 | 4 | 4 | 100 | 4 | 3 | 75 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Fish | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomato
Carrot |
2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| 3 | 2 | 66.7 | 3 | 3 | 100 | 3 | 2 | 66.7 | |
| Total | 27 | 26 | 96.3 | 27 | 27 | 100 | 27 | 25 | 92.3 |
Examples of PCR amplification products from detectable DNA of contaminated food were shown in (Tables 9 and 10).
Table (9):
PCR of Salmonella Typhi for contaminated foods after different incubation periods.
| Type of food | Directly tested without incubation | Tested after 8 hrs incubation | Tests of overnight culture | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 1 | 50 |
| Milk | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Egg | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 4 | 100 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Fish | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomato
Carrot |
2 | 1 | 50 | 2 | 1 | 50 | 2 | 1 | 50 |
| 3 | 3 | 66.7 | 3 | 3 | 100 | 3 | 2 | 66.7 | |
| Total | 27 | 26 | 96.6 | 27 | 26 | 96.3 | 27 | 24 | 88.9 |
Table (10):
PCR of Shigella dysentery for contaminated foods after different incubation periods.
| Type of food | Directly tested without incubation | Tested after 8 hrs Incubation |
Tests of overnight culture | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | +ve | % | No | +ve | % | No | +ve | % | |
| Yogurt | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 1 | 50 |
| Milk | 4 | 4 | 100 | 4 | 4 | 100 | 4 | 3 | 75 |
| Egg | 4 | 3 | 75 | 4 | 3 | 75 | 4 | 4 | 10 |
| Bread | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Meat | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Chicken | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Fish | 2 | 1 | 50 | 2 | 2 | 100 | 2 | 1 | 50 |
| Beef burger | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Sausage | 2 | 2 | 100 | 2 | 2 | 100 | 2 | 2 | 100 |
| Tomato
Carrot |
2 | 2 | 100 | 2 | 2 | 100 | 2 | 1 | 50 |
| 3 | 2 | 66.7 | 3 | 2 | 66.7 | 3 | 2 | 66.7 | |
| Total | 27 | 23 | 85.2 | 27 | 25 | 92.6 | 27 | 22 | 77.8 |
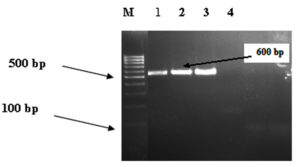
We obtained 1.5% Agarose gel electrophoresis of PCR products from DNA extracted from contaminated food samples. Lanes M, molecular size markers (100 bp ladder); lanes 1, 2,3 positive result; and lane 4, negative Shigella dysentery IpaH gene result as shown in (Fig. 3).
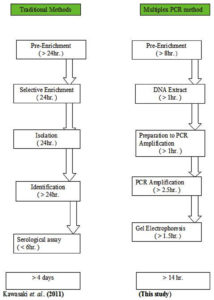
The developed multiplex PCR method for identification of E. coli, Salmonella Typhi, and Shigella dysentery used in this study is showed in (Figure 4). The comparison between traditional and current Multiplex PCR methods for identification of species used in this study is showed in (Figure 5).
Fig. 4. Developed Multiplex PCR method for identification of E. coli, Salmonella Typhi, and Shigella dysentery used in this study.
This study is aimed to facilitate rapid, accurate detection in reasonable efforts and costs that are attainable. A rapid multiplex PCR method was performed to detect Escherichia coli, Salmonella Typhi, and Shigella dysentery strains contaminating food. Two sets of primers for each bacterium were chosen. Of the six sets of PCR primers, the Mdh primer pair for Escherichia coli, IpaB primer pair for Salmonella Typhi, and IpaH1 primer pair for Shigella dysentery were the best primers sets. Many multiplex PCR studies were done before to detect the foodborne pathogens in food, Iun et al.26 identified six common foodborne pathogens. Detection of Escherichia coli O157: H7 using multiplex PCR was developed by Apirak et al.31 The suitability of a PCR procedure was evaluated as a means of detecting Salmonella species by Pathmanathan et al.32 Fukushima et al. established a new phylogenetic tree for the classification of Salmonella, Shigella, and Escherichia coli using the PCR method.32 In this study, the sensitivity to detect E. coli, Salmonella Typhi, and Shigella dysentery in contaminated food in the concentration of the infective and the over infective doses were 100%, 96.3%, and 88.9% respectively for the three bacteria. The sensitivity of the methods was 100% in a beef burger, meat, and bread samples, and then it decreased in other types of foods. In the less than the infectious dose concentration, the sensitivity of E. coli was about 88.9%. In such doze concentrations, the sensitivity of the methods was less effective in some types of foodstuff like yogurt (pH ≤ 4.4) which showed a lower sensitivity of 50% and a sensitivity of 75% for the eggs (pH approximately 8).34 This may be due to large variation in the pH of these types of food that may affect the PCR optimization conditions and exceed the capacity of the buffer used.
The multiplex PCR for the specific detection of Salmonella Typh is showed a sensitivity of (92.6%) and there was no variation in sensitivity between the different contaminated foods. Nevertheless, a lower sensitivity was recorded in yogurt. The sensitivity of the detection of Shigella dysentery in less infected doses was equivalent to 66.7%. Such low sensitivity may attribute to the fewest number of cells in such infectious dose which is only 10 cells.35 Determination of an optimum sampling time for bacterial inspection is a crucial factor for food safety. Tests carried out in the laboratory for contaminated samples showed no significant difference in detection of bacteria after incubation for 8 hours, 24 hours, or even without incubation period. Therefore, the most suitable, reliable, and sensitive result for all three bacteria was obtained without incubation period or after 8 hours of incubation. By the multiplex PCR method used in this study not only the time will decrease to less than 15 hours, but also the assessment of the microbiological quality of food, water, and medications will happen rapidly, resulting in increased safety.
A multiplex PCR technique as a rapid method; was performed using Mdh primer pair to detect Escherichia coli, IpaB primer pair to detect Salmonella Typhi, and IpaH1 primer pair to detect Shigella dysentery. The sensitivity of the mPCR was very high, even in concentrations of less infected doses. The three bacterium species were all detected (100%) in the limit of infected and over infected doses. The most suitable, reliable, and sensitive result for all three bacteria can obtain without incubation period or after 8 hours of incubation.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
EBHA, MOE, and NME performed the experiments. EBHA, MOE, NME, THS, AMA, AAO, and MSA analyzed the data. EBHA, MOE, NME, THS, AMA, AAO, and MSA wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- WHO. WHO Global strategy for food safety: safer food for better health 2010. Geneva: WHO – ISBN 92 4 154574 7.
- WHO – Foodborne disease outbreaks: Guidelines for investigation and control, 2008. Geneva: WHO – ISBN 978 92 4 154722 2.
- CDC-Diagnosis and Management of Foodborne Illnesses – A Primer for Physicians and Other Health Care Professionals – Produced collaboratively by the American Medical Association – American Nurses Association- American Nurses Foundation – CDC, FDA, FSIS, US Department of Agriculture – USA 2004.
- Richard L, Laurie C and Judy D – The Food Safety Hazard Guidebook – 2ndedition – The Royal Society of Chemistry, 2012; p: 10 -104 – ISBN: 978-0-85404-460-3.
- CDC: Centre for Disease Control and Prevention, USA – CDC estimates of Foodborne illness in the United States – National Center for Emerging and Zoonotic Infectious Disease – Division of Foodborne, Waterborne and Environmental Diseases,2012; CS218786-A – www.cdc.gov/foodborneburden.
- Hussein AH. Aerobic Bacteria in Fresh and Refrigerated Beef. Khartoum: M.V.Sc thesis, University of Khartoum. 1975.
- Abd ElRahman AE – Some Aerobic bacteria in minced meat sold in retail in Elmokhtar Municipality. Khartoum: M.Sc thesis, University of Khartoum, 1999.
- Mohamed AE – Aerobic bacteria isolated from meat at different stages of processing. Khartoum: M.V.Sc thesis, University of Khartoum 2000.
- Ahmed EB – Escherichia- coli and Bacterial load in fried Burger Sold at Alarabi Market, Locality of Khartoum – Journal of Art and Sciences – Dallanj University, 2006; 2: 1-26.
- Ahmed EB – Escherichia- coli and Bacterial load in fried Burger Sold at Alarabi Market, Locality of Khartoum – MPEH thesis – Faculty of Public and Environmental Health – Khartoum University, 2003.
- Warsama LM, El Zubeir, IEM and El Owni OAO– Composition and Hygienic Qualityof Sudanese White Soft Cheese in Khartoum North Markets (Sudan) – International Jurnal of Dairy Science, 2010;5 (3): 177-184 – ISBN 1811-1743.
Crossref - Hassan NA – Bacterial Contamination of Restaurant food in Khartoum State: M.V.Sc thesis – University of Khartoum 2006.
- CDC Bulletin – Monitoring the Incidence and Causes of Diseases Potentially Transmitted by Food in Australia: Annual Report of the OzFooDNet, 2009;34(4):396-426.
- Mustafa NEM and MS Abdallah – Bacteriological Quality of Street -Vended Um-Jingir: A Traditional Sudanese Food – African Journal of Food, Agriculture, Nutrition and Development, 2011;11(5).
- Jasson V, Jacxsens L, Luning P, Rajkovic A, Uyttendaele M (2010) – Review. Alternative microbial methods: An overview and selection criteria – Food Microbiol, 27 – p: 710-730.
- Bekal S, Brousseau R, Masson L, Prefontaine G, Fairbrother J and Harel J – Rapid Identification of Escherichia coli Pathotypes by Virulence Gene Detection with DNA Microarrays – Journal of Clinical Microbiology, 2003; 41(5): 2113–2125.
Crossref - Babalola OO – Molecular techniques: An overview of methods for the detection of bacteria – African Journal of Biotechnology, 2003; 2(12): 710-713, December 2003 – ISSN 1684–5315 – Academic Journals Minireview – http://www.academicjournals.org/AJB.
- Wang Y, Zunzhong Y and Ying Y – New Trends in Impedimetric Biosensors for the detection of foodborne pathogenic Bacteria – Sensors, 2012; 12 – p: 3449-3471.
Crossref - Abubakar I, Irvine L, Aldus CF, Wyatt GM, Fordham R, Schelenz S, Shepstone L, Howe A, Peck M, Hunter PR – A systematic review of the clinical, public health and cost-effectiveness of rapid diagnostic tests for the detection and identification of bacterial intestinal pathogens in faeces and food – Health Technol Assess, 2007 11 – p:1–216.
- Dennis YML, Rossa WKC and KC Allen C – Clinical Applications of PCR – 2nd Ed – p : 1-18, 2006. Humana Press Inc. – New Jersey.
- Arthur TM, Bosilevac JM, Nou X and Koohmarie M. Evaluation of culture and PCR based detection methods for E. coliO157:H7 in inoculated ground beef. J. Food Prot.,2005;86:1566-1574.
- John MS, Bartlett and D Stirling. Methods in Molecular Biology, PCR Protocols, 2003; 226 – 2nd Ed – p : 1-18 – Humana Press Inc., Totowa, New Jersey 07512.
- Bai S, Zhao J, Zhang Y, Huang W, Xu &Haodong S, Chen H, Fan L, Chen Y, Deng X.W. Rapid and reliable detection of 11 food-borne pathogens using thin-film biosensor chips- Appl Microbiol Biotechnol, doi: 10.1007/s00253-009-2417-6 – springer.
- Mieta SN, MD Sobsey, TG Barnard – Optimisation of methods for the collection and detection of bacterial pathogens from diarrhoeal human faecal samples using a novel stool collection kit – The 2010 Southern African Young Water Professionals Conference, Pretoria, 19-20 January 2010 – ISSN 0378-4738.
- Jamil M, Bashir S, Mohsin M, et al. Differentiation of common Gram-negative pathogens by PCR-Ribotyping – Pakistan Journal of Medical Science, 23(2):233-237 – www.pjms.com.pk.
- Iun-F, Paul R, Chris B and Ken G – Development of a Multiplex PCR Method for the Detection of Six Common Foodborne Pathogens, Journal of Food and Drug Analysis, 2008;16(4): 37-43.
- Moussa IM, et al., Using molecular techniques for rapid detection of Salmonella serovars in frozen chicken and chicken products collected from Riyadh, Saudi Arabia. African Journal of Biotechnology, 2010; 9(5): 612-619 – ISSN 1684–5315 – http://www.academicjournals.org/AJB.
- Kothary MH and Babu US. Infective Dose of Foodborne Pathogene in Voluntreers: A review – Journal of Food Safety, 2001;21(1): 49–68.
- Lawley R, Curtis L and Davis J. The Food Safety Hazard Guidebook – The Royal Society of Chemistry, 2008 ; Cambridge: 10 -104 – ISBN: 978-0-85404-460-3.
- Gantois I, Ducatelle R, Pasmans F, Haesbrouck F, Gast R. Humphrey TJ, VanImmerseel F. Mechaniosms of egg contamination by Salmonella Enteritidis – FEMS Microbiolo Rev, 2009;33(4):718-738.
- Apirak V, Kobchai P, Jiraporn T, Ryosuke M, Akio K and Orasa S. Detection of Escherichia coli O157: H7 VT and RFBO157 by Multiplex Polymerase Chain Reaction – Correspondence: JirapornThaniyavarn, Department of Microbiology, Faculty of Science, Chulalongkorn University, Bangkok, Thailand., 2007;38:82-90.
- Pathmanathan, S. Cardona-Castro, M. Sa´nchez-Jime´nez, M. Correa-Ochoa, S.Puthucheary and K. Thong. Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene – Journal of Medical Microbiology, 2003;52: 773–776
Crossref - Fukushima M, Kakinuma K, and Kawaguchi R. Phylogenetic Analysis of Salmonella, Shigella, and Escherichia coli Strains on the Basis of the gyrB Gene Sequence, Journal of Clinical Microbiology, 40(8): 2779–2785
Crossref - FDA – Approximate pH of Foods and Food products – FDA/Center for Food Safety and Applied Nutrition, 2003; updated March 24, 2004.
- Okolie N, Omonigehin E, Badru OA and Akande IS. Isolation of pathogenic bacteria from some foods sold at selected private schools in Akoka area of Yaba – Lagos, Nigeria, African Journal of Food Science, 2012;6(3):65-69.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.