ISSN: 0973-7510
E-ISSN: 2581-690X
Leptospirosis is a zoonosis prevalent in tropical countries and affects animals and humans alike. Leptospira interrogans, the causative organism for this waterborne infection, spreads through the urine of infected animals. There is a direct link between contaminated water and Leptospira outbreaks. This study reports a rapid assay to detect and differentiate pathogenic Leptospira from non-pathogenic in environmental water using multiplex PCR. The assay uses primers targeting the Lipl32 and Lipl21 gene. The multiplex PCR has been standardized using 11 pathogenic and one saprophytic serovar of Leptospira. The analytical sensitivity of the developed method was evaluated with different concentrations of template DNA. This method was used to screen water samples collected from 20 different sources from Chengalpattu town in Kancheepuram District, Tamil Nadu, India. Of the 20 water samples screened, 13 samples tested positive for pathogenic Leptospira, and seven samples tested negative. Four water samples were found to carry both pathogenic and saprophytic species. The developed multiplex PCR assay is highly useful for detecting and distinguishing pathogenic and saprophytic leptospires in water.
Leptospirosis, waterborne infection, water, pathogenic serovars, multiplex PCR
Leptospirosis is a zoonosis having a global presence with a higher prevalence in tropical regions. Heavy rainfall and flooding result in large epidemics causing deaths in developed and developing countries1-3. Humans are infected via exposure to the urine of infected animals or through contaminated soil or water 4. Leptospira species display different affinities and adaptations in different hosts and exhibit clinical symptoms of varying degrees5. Rodents are reservoirs and carriers of Leptospira but rarely show symptoms of the disease.
Meanwhile, other animals like livestock, cattle, and pets exhibit severe symptoms, including multiple organ injury and abortion upon infection6,7. Similarly, in infected humans, the disease’s clinical manifestations range from mild, flu-like symptoms to severe conditions like jaundice, renal impairment, and hemorrhage to life-threatening conditions5. A direct correlation between Leptospira epidemics and direct contact with contaminated environmental water has been reported. A damaged skin barrier like a cut or an open wound is the most common route of pathogen entry2,8,9.
Based on 16S rRNA phylogeny, the genus Leptospira is arranged into three large subgroups and includes at least 21 species. Pathogenic (Group I), Intermediate pathogenic (Group II), and saprophytes are the three subgroups. The infectious groups I and II include 14 species, and the non-infectious group III comprises 7 species referred to as saprophytes and is not known to cause infections10. Group I pathogens like L. kirschneri, L. noguchii, and L. interrogans have 250 distinct serotypes. They cause disease whose symptoms range from mild infections to severe disease and death. Intermediate pathogens mostly cause mild illnesses that are easily treatable and without fatal complications. Saprophytic Leptospira belongs to group III. They are free-living environmental microorganisms and are morphologically similar to pathogenic Leptospira, making it hard to discriminate between them by microscopic observation alone11.
Currently, the microscopic agglutination test (MAT) is used to detect leptospirosis. Though this is deemed the gold standard, it lacks sensitivity and cannot be used for early diagnosis. Culture-based methods are time-consuming and hence unsuitable for rapid diagnosis. Several molecular methods, including conventional PCR, duplex PCR, nested PCR, real-time PCR, and loop-mediated isothermal amplification (LAMP), have been used to detect pathogenic Leptospira in clinical samples12-16. These PCR based target virulent genes like Lipl32, lfb, ligA, and ligB217-19. Sequence variations in the housekeeping genes, including gyrB, rrs, and 16S rRNA, have also been exploited to distinguish between pathogenic and saprophytic Leptospira in clinical samples 19,20.
This study is designed to develop a multiplex PCR assay for rapid detection and differentiation of Leptospira strains in environmental water. In order to achieve this, a combination of primers that target a portion of the Lipl21 gene unique to saprophytic Leptospira and pathogen-specific Lipl32 gene were used. Lipl32 codes for a surface lipoprotein with a molecular weight of 32kDa and is present only in pathogenic Leptospira. Meanwhile, Lipl21 encodes another lipoprotein with a molecular weight of 21kDa. The study was performed using 12 Leptospira strains and was successfully used to detect and differentiate between the Leptospira strains in environmental water samples.
Leptospira Strains
A total of 12 Leptospira strains obtained from Dr. Natarajasrinivasan, Department of Microbiology, Bharathidasan University, Tiruchirappalli, Tamil Nadu, India, were used in this study (Table 1). The cultures were maintained in a liquid medium (Leptospira Medium Base M1009, Himedia) supplemented with 5-fluorouracil and Leptospira Enrichment (Himedia). All cultures were incubated at 30°C.
Table (1):
List of Leptospira strains used in this study.
| S.No | Leptospira strains | Lipl21 | Lipl32 |
|---|---|---|---|
| 1 | L. interrogans serovar Icterohaemorrhagiae | – | + |
| 2 | L. interrogans serovar Hebdomadis | – | + |
| 3 | L. interrogans serovar Pomona | – | + |
| 4 | L. interrogans serovar Pyrogens | – | + |
| 5 | L. interrogans serovar Sejroe | – | + |
| 6 | L. interrogans serovar Bataviae | – | + |
| 7 | L. interrogans serovar Autamnalis | – | + |
| 8 | L. interrogans serovar Canicola | – | + |
| 9 | L. interrogans serovar Australis | – | + |
| 10 | L. kirschneri serovar Grippotyphosa | – | + |
| 11 | L. borgpetersenii serovar Ballum | – | + |
| 12 | L. meyeri serovar Semaranga | + | – |
| “-”: absent “+”: present | |||
Primers Used
Oligonucleotide primers used in this study (Table 2) were synthesized by Priority Life Science, Coimbatore, Tamil Nadu, India. These primers were used to amplify Lipl21 (582 bp) and Lipl32 (719 bp) regions from L.meyeri serovar Semarangaa and L.interrogans serovar Icterohaemorrhagiae respectively. The 582 bp region amplified by Lipl21 FP and Lipl21 RP is specific to saprophytic Leptospira21.
Table (2):
List of primers used in this study.
| Target Region | Primer | Sequence (5’-3’) | Reference |
|---|---|---|---|
| Lipl32 | Lipl32 FP | CCCCATGAAAAAACTTTCGATTTTGGC | 21 |
| Lipl32 RP | CCCCTGGAATACCTGGTGGGAAAA | ||
| Lipl21 | Lipl21 FP | ATGAAGAAATCACTTATCG | |
| Lipl21 RP | TTACTGAACAGCAGTTGC |
DNA Extraction
All genomic DNA used in the study was isolated using DNeasy Blood and Tissue kit by Qiagen (Germany). 100 ng of genomic DNA from the different pathogens was used for subsequent PCR reactions.
Sample collection
Twenty different water samples were collected from in and around Chengalpattu town in Kancheepuram District, Tamil Nadu, India. The collected water samples were stored in a sterile glass container at -20°C. The water samples were concentrated for 45 minutes using a vacuum concentrator (Eppendorf, India) and used as templates for multiplex PCR.
Multiplex PCR reaction
A 30 µL PCR mix contains 100 ng of genomic DNA from respective pathogens, 10 pM/L of each primer, 10 mM/L KCl, 20 mM/L Tris–HCl, 8 mM/L MgSO4, 10 mM/L (NH4)2SO4, 1U of Taq DNA polymerase. The amplicons were visualized on a 1.5% agarose gel stained with ethidium bromide. Lipl21 and Lipl32 amplicons were cloned into the pGEM®T-Easy vector (Promega, USA) and used as positive template controls in the study. The multiplex PCR used an initial denaturation temperature of 95°C for 5 minutes, 35 iterations of denaturation (94°C – 30 seconds), annealing (55°C – 45 seconds) and extension (72°C- 45 seconds), followed by a final extension at 72°C for 5 minutes. Analytical sensitivity of the multiplex PCR was evaluated with varying template DNA concentrations (50 ng/µL, 12.5 ng/µL, 125 ng/µL, 780 pg/µL, 190 pg/µL, 47.5 pg/µL). All experiments were performed in triplicates.
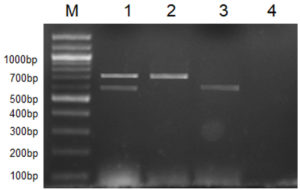
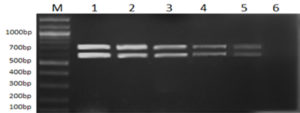
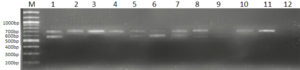
All 12 cultures used in this study were screened for the presence of their respective marker gene (i.e., Lipl32 in the case of pathogenic species and a stretch of DNA sequences in Lipl21 that is found explicitly in non-pathogens). A multiplex PCR technique was developed using recombinant plasmids containing genes Lipl21 and Lipl32. The amplification products of 582 bp (Lipl21) and 719 bp (Lipl32) were observed in a single PCR reaction. The products were confirmed by comparing the uniplex PCR products of Lipl21 and Lipl32 (Fig. 1). The multiplex PCR’s analytical sensitivity was found to be 190 pg/µL (Fig. 2). All the pathogenic cultures used in this study amplified the Lipl32 region except the saprophytic Leptospira serovar Semaranga, which tested positive for Lipl21. None of the cultures showed amplification products of both genes (Fig. 3). These outcomes indicate the specificity of the developed method.
Fig. 1. Multiplex PCR for pathogenic and saprophytic Leptospira. M: 100 bp marker, 1: Lipl32 and Lipl21, 2: Lipl32 from L. interrogans ser. Icterohaemorrhagiae, 3: Lipl21 from L. meyeri ser. Semaranga, 4: Negative control.
Fig. 2. Sensitivity analysis of Multiplex PCR with different template concentrations of Lipl32 and Lipl21. Lane M: 100bp Marker, 1: 50ng/µL, 2: 12.5ng/µL, 3: 3.125ng/µL, 4: 780pg/µL, 5: 190pg/µL, 6: 47.5pg/µL.
Fig. 3. Multiplex PCR with pathogenic and saprophytic cultures
Lane M: 100bp Marker, 1: Lipl32 and Lipl21 plasmid (positive control). 2: L. interrogans ser. Icterohaemorrhagiae, 3: L. interrogans ser. Hebdomadis, 4: L. interrogans ser. Pyrogens, 5: L. interrogans ser. Sejroe, 6: L. kirschneri ser. S Grippotyphosa, 7: L.interrogans ser. Bataviae, 8: L. interrogans ser. Canicola, 9: L. borgpetersenii ser. Ballum, 10: L.interrogans ser. Pomona, 11: L. interrogans ser. Autamnalis, 12: L interrogans ser. Australis, 13: L.meyeri ser. Semarangaa, 14: Negative control.
In this study, the water samples were concentrated and used as a template for a multiplex PCR. Thirteen water samples tested positive for pathogenic Leptospira, and seven samples showed negative amplification for pathogenic and saprophytic Leptospira. Four water samples were found to carry both pathogenic and saprophytic Leptospira. Few amplification products of negative and positive reactions are shown in Fig. 4.
The misdiagnosis of leptospirosis can be attributed to the similarity between the clinical symptoms of leptospirosis and other febrile diseases like dengue and malaria. This, in turn, has led to the underestimation of the prevalence of the disease22. Long incubation periods required for the growth of Leptospira make culturing difficult for diagnosis, and observation using dark-field microscopy is unreliable. The serological test MAT, which is considered the benchmark technique for diagnosing this disease, has several drawbacks as it is arduous, slow, and requires maintaining an elaborate collection of reference strains23,24.
The extensive use of molecular techniques has upgraded the speed, sensitivity, and specificity of leptospirosis diagnosis. Many PCR-based methods have been reported for Leptospira detection, but each technique comes with limitations16. The sensitivity and specificity of the PCR significantly depend on choosing reliable target genes and primer optimization. The marker gene Lipl32 used in this study is specific to pathogenic Leptospira species. The Lipl21 gene is present in both pathogenic and non-pathogenic Leptospira, but sequence variations exist between the Lipl21 sequence of these two Leptospira groups. We exploited this sequence variation and developed a multiplex PCR targeting Lipl32 and Lipl21 genes to screen for pathogenic and saprophytic Leptospira species. When performed using genomic DNA from pathogenic and non-pathogenic cultures, the assay showed an analytical specificity of 100%. Water samples from multiple sources were tested using this multiplex PCR method to determine the assay’s effectiveness in the field. It was observed that the multiplex PCR assay successfully detected & differentiated between the two groups, even in water samples that contained both pathogenic and non-pathogenic Leptospira. These results indicate the usefulness of this method for screening environmental samples. This will help take necessary measures to prevent exposure to Leptospira-contaminated water.
ACKNOWLEDGMENTS
The authors convey their heartfelt thanks to Dr. K Natarajan Seenivasan, Department of Microbiology, Bharathidasan University, Tamil Nadu, for providing Leptospira cultures. We sincerely thank the Indian Council of Medical Research for the funding support towards the execution of the project (Leptos/28/2-13-ECD-I). We also thank the management of SRM Institute of Science and Technology for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
This work was supported by the Indian Council of Medical Research (Leptos/28/2-13-ECD-I).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
Details of the isolates and primer sequences used are included in the form of tables. Results of all experiments performed are included as figures in the manuscript.
- Haake DA, Levett PN. Leptospirosis in humans. Curr Top Microbiol Immunol. 2015;387:65-97.
Crossref - Morgan J, Bornstein SL, Karpati AM, et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis. 2002;34(12):1593-1599.
Crossref - Brockmann S, Piechotowski I, Bock-Hensley O, et al. Outbreak of leptospirosis among triathlon participants in Germany, 2006. BMC Infect Dis. 2010;10:91.
Crossref - Bharti AR, Nally JE, Ricaldi JN, et al. leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3(12):757-771.
Crossref - Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296-326.
Crossref - Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, Vinetz JM. Asymptomatic Renal Colonization of Humans in the Peruvian Amazon by Leptospira. PLoS Negl Trop Dis. 2010;4(2).
Crossref - Ashford DA, Kaiser RM, Spiegel RA, et al. Asymptomatic infection and risk factors for leptospirosis in Nicaragua. Am J Trop Med Hyg. 2000;63(5):249-254.
Crossref - Ciceroni L, Stepan E, Pinto A, et al. Epidemiological trend of human leptospirosis in Italy between 1994 and 1996. Eur J Epidemiol. 2000;16(1):79-86.
Crossref - Kucerova P, Cermakova Z. Leptospirosis: a neglected zoonosis of global distribution. Rev Med Microbiol. 2013;24(3):63-69.
Crossref - Lehmann JS, Matthias MA, Vinetz JM, Fouts DE. Leptospiral Pathogenomics. Pathogens. 2014;3(2):280-308.
Crossref - Vk C, Ty L, Wf L, et al. leptospirosis in human: Biomarkers in host immune responses. Microbiol Res. 2018;207:108-115.
Crossref - Benacer D, Zain SNM, Lewis JW, et al. A duplex endpoint PCR assay for rapid detection and differentiation of Leptospira strains. Rev Soc Bras Med Tro. 2017;50(2):239-242.
Crossref - Blanco RM, Romero EC. Evaluation of nested polymerase chain reaction for the early detection of Leptospira spp. DNA in serum samples from patients with leptospirosis. Diagn Microbiol Infect Dis. 2014;78(4):343-346.
Crossref - Merien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992;30(9):2219-2224.
Crossref - Sengupta M, Prabhakar AKP, Satyendra S, et al. Utility of Loop-mediated Isothermal Amplification Assay, Polymerase Chain Reaction, and ELISA for Diagnosis of Leptospirosis in South Indian Patients. J Glob Infect Dis. 2017;9(1):3-7.
Crossref - Waggoner JJ, Balassiano I, Abeynayake J, et al. Sensitive Real-Time PCR Detection of Pathogenic Leptospira spp. and a Comparison of Nucleic Acid Amplification Methods for the Diagnosis of Leptospirosis. PLoS One. 2014;9(11).
Crossref - Figueira CP, Croda J, Choy HA, et al. Heterologous expression of pathogen-specific genes ligA and ligB in the saprophyte Leptospira biflexaconfers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 2011;11(1):129.
Crossref - Perez J, Goarant C. Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. BMC Microbiol. 2010;10:325.
Crossref - Thaipadunpanit J, Chierakul W, Wuthiekanun V, et al. Diagnostic Accuracy of Real-Time PCR Assays Targeting 16S rRNA and Lipl32 Genes for Human Leptospirosis in Thailand: A Case-Control Study. PLoS ONE. 2011;6(1):e16236.
Crossref - Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006;6:95.
Crossref - Monica NI, Rathinasabapathi P, Ramya M. Development of real-time loop-mediated isothermal amplification (RealAmp) method for sensitive and rapid detection of pathogenic and non-pathogenic Leptospira. Lett Appl Microbiol. 2019;68(2):196-203.
Crossref - Benacer D, Mohd Zain SN, Sim SZ, et al. Determination of Leptospira borgpetersenii serovar Javanica and Leptospira interrogans serovar Bataviae as the persistent Leptospira serovars circulating in the urban rat populations in Peninsular Malaysia. Parasite Vector. 2016;9(1):117.
Crossref - Chirathaworn C, Inwattana R, Poovorawan Y, Suwancharoen D. Interpretation of microscopic agglutination test for leptospirosis diagnosis and seroprevalence. Asian Pac J Trop Bio. 2014;4(Suppl 1):S162-S164.
Crossref - Ahmad SN, Shah S, Ahmad FM. Laboratory diagnosis of leptospirosis. J Postgrad Med. 2005; 51(3):195-200.
© The Author(s) 2021 Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.