ISSN: 0973-7510
E-ISSN: 2581-690X
Highly pathogenic avian influenza viruses are circulating in lots of avian species, causing major outbreaks in both wild and domestic poultry. Since its first emergence in 2014, clade 2.3.4.4 H5N8 viruses widely spread in the world resulting in enormous economic losses. In Egypt, the newly emerging high pathogenic avian influenza (HPAI) H5N8 viruses have been detected in domestic poultry and in wild birds since the 2016/2017 winter season. AI H5N8 is cocirculating with LP H9N2 and HP H5N1 in the Egyptian environment. Poultry vaccination strategy in Egypt is based on commercially available H5 vaccines as an essential control policy, while the majority of commercial avian influenza H5 vaccines utilized in Egypt are not effective against H5N8 viruses. The present study included 3 experimental H5N8 inactivated vaccines based on the 2 major antigenic proteins of the currently circulating strain A/chicken/Egypt/Q16684C/2019 (H5N8), and the internal segments of the A/PR/8/1934 (H1N1) virus. Then, the protective efficacy of the three forms of inactivated vaccines (HAH5N8+7PR8, NAH5N8+7PR8 and HA, NAH5N8+6PR8) were compared regarding the parental PR8 virus in vaccinated specific pathogen free chickens. The NAH5N8+6PR8 as well as HAH5N8+7PR8 and HA vaccines showed the highest protection capacity of challenged SPF chickens and were able to elicit the highest titers of virus-neutralizing antibodies. Thus, a continuous active surveillance strategy is needed to determine the most dominant circulating strain and updating of vaccine seed strains.
Avian Influenza Viruses, Egypt, H5N8, Pathogenicity, Vaccine
Avian influenza viruses (AIV) are highly infectious viral agents spreading to several domestic and wild bird species.1 AIV exists in nature in high pathogenic (HP) or low pathogenic (LP) forms, which were classified according to their molecular characteristics and their ability to cause the death of infected chickens in the laboratory.2 Wild birds are proven to be the key source and distributor of influenza A viruses. The wild aquatic birds are considered the main reservoir that harbor’s various subtypes of influenza viruses.3 Wild birds also play a key role in virus transmission and spreading as some wild-type species show little or no signs when infected with influenza viruses, even the highly pathogenic influenza viruses that cause the death of infected domestic chickens.4 This makes the control of influenza viruses transmission is nearly impossible, as the transmission of influenza viruses from infected wild birds to domestic poultry causes the start of new outbreaks in that site, and can need further emerge and reassort with previously emerged circulating viruses. 5,6
One of the newly emerging highly pathogenic viruses is the highly pathogenic H5N8 virus clade 2.3.4, which was first detected in 2014, circulating in Asia, China, and Korea, and then transmitted to other countries in the world through infected migratory wild birds infecting the domestic poultry. From that point, H5N8 was emerging and kept evolving to new subclades of AIVs.7 Highly pathogenic avian Influenza HPAI H5N8 viruses are divided into A and B subgroups. In contrast of group A, which is no longer being detected, group B has emerged worldwide and continued to spread and develop new reassorting forms mainly driven by wild migratory birds. Human health face great threat resulting from the evolution of HPAI viruses which causes major losses for the poultry industry because of chickens’ death. This describes the insistent need to produce a universal flu vaccine that can stimulate sufficient immune response and long-lasting protection against multiple types of influenza viruses.8,9
Three subtypes of AIV are co-circulating in Egyptian farms, HP H5N1 clade 2.2.1 circulating since 2006, LP H9N2 G1 viruses since 2011, and highly pathogenic H5N8 viruses clade 2.3.4 since 2016. The co-circulation of these 3 viruses poses a major threat, which shows that despite the massive efforts exerted in the field of poultry vaccination and controlling the influenza virus, the currently used strategy is not enough and needs to be evolved.
The most predominant control strategy for H5 viruses in Egypt is through vaccination using commercial AI/H5 vaccines. Many vaccines are commercially available to different clades of H5 viruses. Many of these vaccines are available in Egyptian markets. Around 22 different types of inactivated vaccines against H5 viruses are imported in Egypt and used in the market to control virus spreading and to protect chickens from influenza infection and death.
However, the detection and emergence of clade 2.3.4.4 H5N8 viruses in poultry in 2017 in Egypt mean that the currently used commercial H5 vaccine in Egypt is not effective, and the control measures need to be readdressed to protect poultry against the newly emerging H5N8 virus.10
Also, the effectiveness of these commercially available vaccines against H5N8 viruses circulating in Egypt was tested and most of them indicated low efficacy in protection against H5N8 infection.11 Inactivated vaccines are the most commercially available form of influenza vaccines.
This raised the urge to develop new vaccines that can induce long-lasting immune responses and protect against currently circulating viruses and help pursuing the aim of producing the universal influenza vaccine later in the future. The most predominant vaccination strategy used in mass vaccination in the poultry industry is the inactivated influenza vaccine form, especially to control the HP H5 viruses. Inactivated vaccines are formulated through the attenuation of the hemagglutinin of the HPAI parental virus through deleting the cleavage site multi-basic amino acids. The new altered HA is then utilized, with the neuraminidase NA of the same parent circulating virus, plus the six internal segments of a low pathogenic virus (PR/8 H1N1 influenza virus), to generate a new low pathogenic virus as a vaccine candidate by reverse genetics.12
In the present study, the aim is to generate 3 candidate reassortment vaccine strains based on using one or both of the main antigenic proteins in influenza virus (HA and/or NA) gene segments of the currently circulating Egyptian H5N8 A/chicken/Egypt/Q16684C/2019 (H5N8), along with the internal gene segments of the H1N1 A/Puerto Rico/8/34 (PR8) as a backbone. the protection efficacy of the prepared vaccines (HAH5N8+7PR8, NAH5N8+7PR8, and HA, NAH5N8+6PR8) was then compared to the parental PR8.
Ethical approval
Infection experiment was achieved in a biosafety level 3 negative-pressure chicken isolators (Plas Laboratorys, Lansing, MI, USA). Humane endpoint was assigned and culling of animals was performed in case any chicken showed a rapid onset of loss of body weight, unwillingness to feed, disorientation, paralysis, or lethargy. Ethical approval was granted from the National Research Center Ethics Committee under registration number 18040. Specific pathogen free embryonated chicken eggs and 4 weeks old SPF chickens were purchased from Kom Oshim Project; Fayum; Egypt.
AI H5N8 Virus
AI H5N8 Viruses were collected through cloacal swabs from dead chickens in a farm in Qalubia Governorate, Egypt, then all collected samples were extracted using viral RNA extraction through Thermo Scientific GeneJET RNA Purification Kit according to the manufacturer protocol and then tested for influenza virus by RT-PCR of M gene using QIAGEN One-Step RT-PCR Kit. Used primers for influenza confirmations are as follows: (forward: ATGAGYCTTYTAACCGAGGTCGAAACG and reverse: TGGACAAANCGTCTACGCTGCAG) as stated in the,13 then positive influenza samples were selected and HA and NA subtypes were identified by subtyping by PCR,14 and then the virus was then identified as H5N8.
Propagated virus sample was then purified by plaque purification assay and propagated in the allantoic cavity of the specific pathogen free SPF egg; 11 days old. Infected eggs (each virus into 5 SPF eggs) were incubated for 48 hours and then chilled at 4°C for 4 hours prior to virus harvest. Harvested virus was then subjected to RNA extraction using a thermo gen get viral extract kit (Qiagen QIAamp Viral RNA Kit). Purified RNA was then used to amplify the full HA and NA of the H5N8 virus using one step verso kit (QIAGEN One-Step RT-PCR Kit according to manufacturer protocol).HA was amplified as 2 fragments to remove the multi-basic amino acids from the cleavage site and turn the HA from LP form (PLREKRRKR/LF) to (ETR/GLF). as described by Kandeil et al.11.
After sequence verification, amplified HA and NA genes were digested using BsmBI and BsaI enzymes, respectively, and then ligated to a digested pHW2000 plasmid. pHW2000 plasmids harboring HA or NA genes were then transformed into DH5a E. coli cells (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and then subjected to plasmid purification using a plasmid extraction kit (QIAprep Spin Miniprep Kit-QIAGEN) and then confirmed by sequencing. Plasmids were then used to produce vaccine seed strain viruses using the reverse genetic technique as previously described by,12 using PR8 6 internal segments cloned in pHW2000 plasmids, kindly provided by Richard Webby, St. Jude Children’s Research Hospital, Memphis, USA, through Materials Transfer Agreement.
Vaccine Preparation by Reverse Genetics
Two types of cells were used in these reverse genetics to produce the vaccine seed strain viruses, Madin Darby Canine Kidney (MDCK) cells, and 293T cells. Media used for both types of cells were supplemented with 1% antibiotic and 5% FBS. The co-culture of MDCK and 293T cells was prepared using Opti-MEM medium (Gibco, Thermo Fisher Scientific) in ratio 1:3, respectively. After 24 h, constructs were mixed to generate respective recombinant viruses as previously described by,12 and then transfected with the 8 segments of each constellating of seed strains. Transfection harvest was infected in allantoic fluid of SPF egg for the propagation of the full PR8 virus as control and the 3 constellations in present experiment ∆HA of H5N8 + 7 PR8 (HAH5N8+7PR8), NA of H5N8 + 7 PR8 (NAH5N8+7PR8) and HA, NA from H5N8 as control (HA, NAH5N8+6PR8), along with the control PR8 virus. After propagation of virus seed strains and titration by HA assay, all viruses were set to 128 HA units and inactivated using 0.1% formalin. To confirm virus inactivation, 5 SPF eggs were inoculated via the allantoic cavity and were incubated for 72 hours post infection. inactivated antigens were mixed with Montanide ISA 71 VG (Seppic, Courbevoie, France) using 30% of viral antigen and 70% of the adjuvant.
Animals’ vaccination
A total of 50 SPF chickens were obtained one week prior to vaccination and serum samples were collected to test for presence of maternal immunity. Each of the 4 vaccines -that were prepared by reverse genetics – was then used to vaccinate a group of 10 healthy SPF Lohmann white chickens (4-weeks-old) by injection. An additional group of 10 non-vaccinated chickens received (PBS + Adjuvant), was used as control non-vaccinated animals, each was vaccinated using 0.5 ml of the corresponding vaccine. A total 50 SPF chickens were then monitored for 4 weeks post-vaccination and serum samples were collected every week. Serum samples were then tested for the anti H5N8 response using the HI assay.
The first 25 µl of serum samples were treated with three folds of receptor destroy enzyme (RDE II, Denka Seiken, Japan, 75 µL) and incubated overnight at 37°C, then inactivated by incubation for 30 minutes at 65°C. Log2 serial dilutions of treated serum were prepared and incubated with the parental H5N8 virus at conc. Of 4 HA units for 30 minutes. 50µl of 0.5% chicken RBCs were added and incubated with serum virus mix for 30 minutes.
Challenge infection
Wild parental HP H5N8 was tittered using EID50 assay by preparing log10 serial dilution of the virus and then infecting the allantoic cavity of SPF eggs. viral titer was then calculated using Reed and Muench equation. At 5 weeks post-vaccination chickens were then infected with 106 EID50 / 100ml through natural routes (intraocular, intranasal, and intratracheal) and monitored for 10 days. Mortality was recorded every day and oral samples were collected at day 3 post-infection and tittered using EID50 assay.
Infection experiment was achieved in a biosafety level 3 negative-pressure chicken isolators (Plas Laboratorys, Lansing, MI, USA).
Statistical Analysis
GraphPad Prism V5 (GraphPad Inc., San Diego, CA, USA) software was utilized for statistical assessment, using the one-way ANOVA test, and Bonferroni post hoc testing. P < 0.05 were considered statistically significant.
Virus Rescue by reverse genetics
Two gene segments (HA and NA) of HPAI A/chicken/Egypt/Q16684C/2019 (H5N8) virus, clade 2.3.4.4, were successfully amplified and cloned in pHW2000. The HA of the HP A/chicken/Egypt/Q16684C/2019 (H5N8) virus which harbors the sequence of multi basic amino acids at the cleavage site (PLREKRRKR/GLF) was modified into a monobasic structure (ETR/GLF). In order to investigate the protection capacity of the major antigenic proteins of the H5N8 virus, we generated the HA, NAH5N8+6PR8 virus commonly used vaccine form using HA and NA of currently circulating H5N8 virus in Egypt and six segments of PR8 virus (Figure 1). Also, PR8 backbone-based vaccines harboring each of the two gene segments of the antigenic proteins of the H5N8 strain (HAH5N8+7PR8 and NAH5N8+7PR8) were rescued. Rescued reassortant viruses (HA, NAH5N8+6PR8, HAH5N8+7PR8, and NAH5N8+7PR8) and PR8 were propagated for two passages and titrated by hemagglutination (HA) assay. The HA units of the vaccine seed strain viruses were set to 128 HA units per 50µl using phosphate-buffered saline.
Figure 1. Summary of the plasmids used for the generation of rescued viruses by reverse genetics. Blue rectangles indicate gene segments of the PR8 virus, orange rectangles indicate gene segments derived from A/chicken/Egypt/Q16684C/2019 (H5N8) virus.
Inactivation of prepared vaccines were tested by inoculated in the allantoic flied of the specific pathogen free SPF egg, and then tested by HA titer assay. HA assay result were negative that confirmed complete inactivation of prepared influenza vaccine. And embryos of inoculated eggs were a live and showed no mortality at 3 days after inoculation.
Immunogenicity and Protection Capacity of Each Form of Inactivated Vaccines
Serum samples were collected from 10 chickens four weeks old at the start of the experiment. Samples were then tested for the existence of the maternal protection against the 2 original H5N8 and PR8 H1N1 viruses using hemagglutination inhibition (HI) assay. All tested chickens showed complete absence of any antibody protection at the beginning of the vaccination experiment. Chickens were then vaccinated with their corresponding vaccine with observation and follow up for 5 weeks post vaccination. All chickens were healthy and showed no signs compared to the control non vaccinated.
Immunological Reactivity
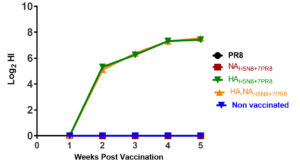
At each week post-vaccination, serum samples were collected and exposed to HI assay versus the parental HPAI H5N8 virus. Only vaccinated chickens with either HAH5N8+7PR8 or HA, NAH5N8+6PR8 specific antibody response detected using HI assay. Antibody response was first detected at 2 weeks post-vaccination (wpv). The antibody titers in the collected serum increased with time up to 4 WPV in the two groups of vaccinated chickens containing HA of H5N8. PR8 and NAH5N8+7PR8 vaccinated groups remained with no anti-H5N8 antibody response based on HI results until week 4 post-vaccination (Figure 2).
Figure 2. The hemagglutination inhibition of the collected serum samples from different vaccination groups at each week post-vaccination against the wildtype HP H5N8 parental virus. The stars represent the significance of the HI titer of HA, NAH5N8+6PR8, and HAH5N8+7PR8 vaccination groups compared to the other three groups at four and five weeks post-infection.
Survival Rate and Virus Shedding
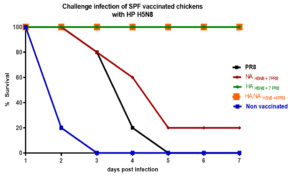
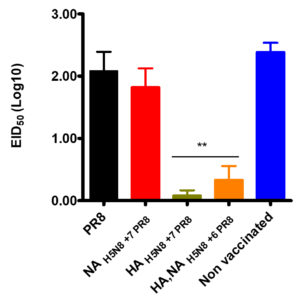
Five chickens from each group were infected with the parental HPAI H5N8 virus, and virus shedding was titrated in oral swabs using EID50. HA, NAH5N8+6PR8 and HAH5N8+7PR8 vaccinated groups indicated a zero death rate and less than 0.5 log10 EID50 viral shedding in swab samples. While PR8 vaccinated group and the non-vaccinated group had no survival as shown in (Figure 3). NAH5N8+7PR8 vaccinated chickens indicated only 20% survival.
Figure 3. The percentage survival rate of the highly pathogenic H5N8 challenged vaccinated and non-vaccinated groups at different days post-infection.
Viral shedding in infected vaccinated groups (HA, NAH5N8+6PR8, and HAH5N8+7PR8) was significantly lesser than all other groups in oral swabs at three days post-infection (Figure 4). No significant differences were observed among NAH5N8+7PR8, PR8, and non vaccinated control groups (p > 0.05).
Emerging of HPAIV poses a continuous threat to animal and public health. New reassortant HPAI H5N8 (HA clade 2.3.4.4) viruses were detected in domestic and wild birds in South Korea, China, and Japan since early 2014,15 followed by several outbreaks in numerous countries in Europe, North America, and Asia is detected in poultry farms and various species of wild birds.16,17 HPAI H5N8 virus was revealed in wild birds and household poultry in Egypt in the 2016/2017 season.18 Genetic characterization indicated that HP H5N8 viruses belong to clade 2.2.3.4 group B, cocirculating in Egyptian farms along with both LP H9N2 and HP H5N1. Co-circulation and co-infection of household poultry may lead to reassortment events and the production of progeny viruses of unknown characteristics and impact on human and animal health. This demonstrates the insistent need to produce a universal flu vaccine that can stimulate sufficient immune response and long-lasting protection against multiple types of influenza viruses.
The most prevalent measure for controlling the HP H5 avian influenza in Egypt is vaccination using commercial avian influenza H5 vaccines. However, most of these commercial vaccines in Egyptian markets previously used to control HP H5N1 were proven inefficient in protecting chickens against the newly emerging H5N8 virus due to the lack of genetic homogeneity between these vaccines and the circulating strain.11 This revealed the need to develop new vaccines based on the strain circulating in Egypt to protect poultry against the newly emerging H5N8 virus.
Thus, in the present study, we intended to determine the role of the two major antigenic proteins of H5N8 virus A/chicken/Egypt/Q16684C/2019 (H5N8) virus in vaccination and protection capacity against the infection with the original wildtype virus in poultry. More studies confirmed that vaccines that have hemagglutinin protein (HAH5N8+6PR8 and HA, NAH5N8+6PR8) produce protection and neutralizing antibodies against the homologous virus, and immunization with inactivated vaccines harboring only the HA glycoprotein induces a strong immunity and full protection after the infection. NAH5N8+6PR8 vaccination indicated low protection capacity (less than 30%.19,20 Similarly, the vaccine including both HA and NA (HA, NAH5N8+6PR8) segments were protective in our experiments, while the NAH5N8+7PR8 provided only 20% protection.
Previous studies revealed a connection between serological reaction and protection against mortality and viral shedding.21 Also, Kumar et. al. reported that low levels of antibody titers could stop mortality, however, can not stop viral shedding. Higher titers of antibodies could prevent mortality and lower viral shedding.22 Former data revealed that serologic titers are linked to the protection when the challenge and vaccine viruses are genetically and antigenically closely related. The presence of HI antibodies predicted protection in the field as well.23 In the infection challenge experiment, HA vaccinated chickens survived while unvaccinated chickens, faced morbidity and mortality.
In conclusion, our data indicated that using the major antigenic glycoprotein gene (HA segment) is critical for effective protection against challenges with genetically homologous viruses, and we recommend the continuous updating of poultry influenza vaccines based on circulating viruses for more effective control and to avoid the viral evolution and formation of escape mutants.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ARS, AAM, AK participated in the design of the study. KEl, ANElT, YM, MElS did all the experimental procedures. KEl, AK, YM, ANElT participated in writing and reviewing the manuscript. And data analysis. AK and MAA reviewed and acquired funding.
FUNDING
This work was funded by the National Research Centre in Egypt, under contract number 12010125.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by National Research Center Ethics Committee under registration number 18040.
- Kayali G, Kandeil A, El-Shesheny R, et al. Avian Influenza A(H5N1) Virus in Egypt. Emerg Infect Dis. 2016;22(3):379-388.
Crossref - CDC. Avian Influenza in Birds. 2017. https://www.cdc.gov/flu/avianflu/avian-in-birds.htm
- Venkatesh D, Poen MJ, Bestebroer TM, et al. Avian Influenza Viruses in Wild Birds: Virus Evolution in a Multihost Ecosystem. J Virol. 2018;92(15):e00433.
Crossref - CDC. Avian Influenza in Birds. 2022. https://www.cdc.gov/flu/avianflu/avian-in-birds.htm
- Shortridge KF. The next pandemic influenza virus? Lancet. 1995;346(8984):1210-1212.
Crossref - Webster RG, Peiris M, Chen H, Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12(1):3-8.
Crossref - de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. A pandemic warning? Nature. 1997;389(6651):554.
Crossref - El-Shesheny R, Barman S, Feeroz MM, et al. Genesis of Influenza A(H5N8) Viruses. Emerg Infect Dis. 2017;23(8):1368-1371.
Crossref - Kang HM, Lee EK, Song BM, et al. Novel reassortant influenza A(H5N8) viruses among inoculated domestic and wild ducks, South Korea, 2014. Emerg Infect Dis. 2015;21(2):298-304.
Crossref - Kandeil A, Hicks JT, Young SG, et al. Active surveillance and genetic evolution of avian influenza viruses in Egypt, 2016-2018. Emerg Microbes Infect. 2019;8(1):1370-1382.
Crossref - Kandeil A, Sabir JSM, Abdelaal A, et al. Efficacy of commercial vaccines against newly emerging avian influenza H5N8 virus in Egypt. Sci Rep. 2018;8(1):9697.
Crossref - Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97(11):6108-6113.
Crossref - WHO information for molecular diagnosis of influenza virus samples – update 2011. https://www.who.int/influenza/resources/documents/molecular_diagnosis_influenza_virus_humans_update_201108.pdf
- Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods. 2001;97(1-2):13-22.
Crossref - Claes F, Morzaria SP, Donis RO. Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr Opin Virol. 2016;16:158-163.
Crossref - Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. J Virol. 2015;89(12):6521-6524.
Crossref - Verhagen JH, Herfst S, Fouchier RA. Infectious disease. How a virus travels the world. Science. 2015;347(6222):616-617.
Crossref - Kandeil A, Kayed A, Moatasim Y, et al. Genetic characterization of highly pathogenic avian influenza A H5N8 viruses isolated from wild birds in Egypt. J Gen Vir. 2017;98(7):1573-1586.
Crossref - Nayak B, Kumar S, DiNapoli JM, et al. Contributions of the avian influenza virus HA, NA, and M2 surface proteins to the induction of neutralizing antibodies and protective immunity. J Virol. 2010;84(5):2408-2420.
Crossref - Moatasim Y, Kandeil A, Mostafa A, et al. Impact of Individual Viral Gene Segments from Influenza A/H5N8 Virus on the Protective Efficacy of Inactivated Subtype-Specific Influenza Vaccine. Pathogens. 2021;10(3):368.
Crossref - Swayne DE, Beck JR, Garcia M, Stone HD. Influence of virus strain and antigen mass on efficacy of H5 avian influenza inactivated vaccines. Avian Pathol. 1999;28(3):245-255.
Crossref - Kumar M, Chu HJ, Rodenberg J, Krauss S, Webster RG. Association of serologic and protective responses of avian influenza vaccines in chickens. Avian Dis. 2007;51(1 Suppl):481-483.
Crossref - Swayne DE, Suarez DL, Spackman E, et al. Antibody titer has positive predictive value for vaccine protection against challenge with natural antigenic-drift variants of H5N1 high-pathogenicity avian influenza viruses from Indonesia. J Virol. 2015;89(7):3746-3762.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.