ISSN: 0973-7510
E-ISSN: 2581-690X
The COVID-19 pandemic caused by the SARS-CoV-2 virus has posed a global challenge. Experts from various branches of science have endeavoured to find solutions to control its spread, one of which has been the quick and precise detection of the virus and its variants in patients. This study aimed to detect the presence of SARS-CoV-2, notably the rapidly spreading Omicron variant, using the spike (S)-gene target failure (SGTF) and S-gene target positive (SGTP) with the principle of the single nucleotide polymorphism (SNP)-probe test. Our descriptive experimental approach detected Omicron variants with the SNP-probe technique using samples of SARS-CoV-2 patients and controls. The probes were designed to recognize the nucleotide code of the amino acids in positions 371 and 417 of SARS-CoV-2. The existence of variants was monitored by the presence or absence of a fluorescence signal, which was translated into a sigmoidal graph using a real-time (RT)-PCR machine. One hundred and twelve samples that had tested positive for SARS-CoV-2 and the Omicron variant using a registered commercial kit showed a similar result to our in-house-developed SNP-probe 371 and 417 assays. The results of this study indicate that the SNP-probe we designed can be used in the detection of the SARS-CoV-2 Omicron variant.
Coronavirus, COVID-19, Omicron Variant, RT-PCR, SARS-CoV-2, SGTF, SGTP, SNP-probe
COVID-19 caused by the SARS-CoV-2 virus has become a worldwide problem. In Indonesia, the first three cases were reported in March 2020. Cases numbers rose slowly until January 2021, then fell from February to June 2021. They increased again in July 2021, reaching almost 50,000 cases, and then from July 2021 to early 2022 to reach almost 60,000 new cases in total.1-3
Changes in the genome of the virus have been reported, resulting in several SARS-CoV-2 variants. The detection of SARS-CoV-2 variants is necessary to monitor and follow the development of COVID-19 cases; however, whole-genome sequencing (WGS) is both costly and time-consuming.4,5 Based on WGS examination, the Indonesian Ministry of Health reported that the rise in case numbers during the period July to October 2021 was caused by the Delta variant of SARS-CoV-2, while the cases from January to February 2022 were caused by the Omicron variant.6
WGS examination can be performed in only a few laboratories in Indonesia as it requires expensive equipment and adequate infrastructure. Our institution, as one of the referral laboratories for COVID-19 diagnosis in Banten province, only conducts WGS in certain cases due to the expensive and complicated process. The results of all cases were reported either to the Indonesian Ministry of Health or the Global Initiative on Sharing All Influenza Data (GISAID) website. Therefore, it is necessary to develop a simpler method for detecting SARS-CoV-2 variants as accurately as commercial kits and that can examine many samples in a short time.
A single nucleotide polymorphism (SNP)-probe can be used as an alternative to detect variants in an organism. Through this method, we can identify different types of variants of the same species based on changes in one (or two) nucleotide bases of the target gene. Many SNP-probe methods have been developed, both with probe hybridization and hydrolysis, and these methods are useful in many applications for the identification of conditions occurring in an organism.7-9
In early 2020, the first SARS-CoV-2 sequencing was reported as the ancestral Wuhan sequence. Over time, more SARS-CoV-2 sequences have been reported either through GISAID or otherwise, enabling WHO to classify the class of SARS-CoV-2 variants.10-12
Based on our results of sequencing the SARS-CoV-2 genome and a review of the GISAID literature, we developed a screening method for the Omicron variant. Because of its ability to spread quickly, a fast and accurate detection method was needed to assist in collecting data on Omicron COVID-19 cases.
Primer, Probe, g-Block Design
Before identifying the Omicron variant from our samples, we designed the primer, probe and g-block, specific for the spike (S)-gene targeting amino acids 371 and 417. Referring to GISAID and the next genome sequencing (NGS) results, we grouped frequently encountered or reported amino acid changes. Table 1 shows the groupings of amino acid changes in each variant, from which amino acid sequence we reversed to the nucleotide sequences. The latter was used to design the primers, probes, and g-blocks. Primers and probes were selected according to the criteria for their requirements. For SARS-CoV-2 primers targeting the S-gene, we selected areas of the nucleotide sequence unchanged for all variants, either ancestral Wuhan or the Alpha to Omicron variants. For the probes, we performed a region-specific selection of amino acids 371 and 417; both Wuhan types were used for S-gene target failure (SGTF) while Omicron was used for S-gene target positive (SGTP).
Table (1):
Primer and Probe Sequences.
| Primer 371 | Forward | 5′- TGG AAC AGG AAG AGA ATC AGC A -3′ |
| Reverse | 5′- AGT AGG AGA CAC TCC ATA ACA CT -3′ | |
| Probe SGTF 371 | Probe 1 | 5′- TCC GCA TCA TTT TCC ACT TT -3′ |
| Probe SGTP 371 | Probe 1 | 5′- CTC GCA CCA TTT TTC ACT TT -3′ |
| Primer 417 | Forward | 5′- TCA GAC AAA TCG CTC CAG GG -3′ |
| Reverse | 5′- CAA GCT ATA ACG CAG CCT GT -3′ | |
| Probe SGTF 417 | Probe 2 | 5′- AAG ATT GCT GAT TAT AAT TA -3′ |
| Probe SGTP 417 | Probe 2 | 5′- AAT ATT GCT GAT TAT AAT TA -3′ |
We used g-blocks as positive or negative controls for both Wuhan and Omicron, SGTF or SGTP, respectively. We also used NGS-positive samples for comparison. In SGTF, the probe attachment to the Wuhan g-block or the Wuhan-NGS variant control sample served as a negative control, while neither attachment to the Omicron g-block nor the Omicron-NGS control sample served as a positive control, and vice versa with SGTP. The primer-probe sequences are listed in Table 1 and the g-block sequences are shown in Table 2.
Table (2):
g-Block Sequences for Wuhan and Omicron.
g-BlockType/ Variant |
Nucleotide Sequence |
|---|---|
Wuhan |
CGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGT AATGATCCATTTTTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTAT TCTAGTGCGAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTA ATTTCAAAAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTA TTAATTTAGTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAAC ATCACTAGGTTTCAAACTTTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGAC AGCTGGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGGACTTTTCTATTAAAATATAATGAAAATGGAA CCATTACAGATGCTGTAGACTGTGCACTTGACCCTCTCTCAGAAACAAAGTGTACGTTGAAATCCTTCACTGTA GAAAAAGGAATCTATCAAACTTCTAACTTTAGAGTCCAACCAACAGAATCTATTGTTAGATTTCCTAATATTAC AAACTTGTGCCCTTTTGGTGAAGTTTTTAACGCCACCAGATTTGCATCTGTTTATGCTTGGAACAGGAAGAG AATCAGCAACTGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCATTTTCCACTTTTAAGTGTTATGGAGT GTCTCCTACTAAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTTGTAATTAGAGGTGATGAAGT CAGACAAATCGCTCCAGGGCAAACTGGAAAGATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGC TGCGTTATAGCTTGGAATTCTAACAATCTTGATTCTAAGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTA GGAAGTCTAATCTCAAACCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAA TGGTGTTGAAGGTTTTAATTGTTACTTTCCTTTACAATCATATGGTTTCCAACCCACTAATGGTGTTGGTTACC AACCATACAGAGTAGTAGTACTTT |
Omicron |
CGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTA ATGATCCATTTTTGGATGTTTATCACCACAAAAACAACAAAAGTTGGATGGAAAGTGGAGTTTATTCTAGTGC GAATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTAATTTCAAA AATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTATTAATTTA GTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTAACATCACTAG GTTTCAAACTTTACTTGCTTTACATACAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCTGGTG CTGCAGCTTATTATGTGGGTTATCTTCAACCTAGGACTTTTCTATTAAAATATAATGAAAATGGAACCATTACAG ATGCTGTAGACTGTGCACTTGACCCTCTCTCAGAAACAAAGTGTACGTTGAAATCCTTCACTGTAGAAAAAGG AATCTATCAAACTTCTAACTTTAGAGTCCAACCAACAGAATCTATTGTTAGATTTCCTAATATTACAAACTTGTG CCCTTTTGGTGAAGTTTTTAACGCCACCAGATTTGCATCTGTTTATGCTTGGAACAGGAAGAGAATCAGCAA CTGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCATTTTCCACTTTTAAGTGTTATGGAGTGTCTCCTACT AAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTTGTAATTAGAGGTGATGAAGTCAGACAAAT CGCTCCAGGGCAAACTGGAAATATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAG CTTGGAATTCTAACAATCTTGATTCTAAGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTCTA ATCTCAAACCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAATGGTGTTGA AGGTTTTAATTGTTACTTTCCTTTACAATCATATGGTTTCCAACCCACTAATGGTGTTGGTTACCAACCATACAGA GTAGTAGTACTTT |
qPCR Analysis
A total of 112 samples from COVID-19 patients who returned a positive SARS-CoV-2 test using the Standard M-nCoV M-NCOV-01 (BioSensor, Korea) were tested for variant confirmation with the SNPsig®-SARS-CoV-2-EscapePLEX (Primerdesign, UK) and VarScreen-RXReady® mBioCoV-19 (BioFarma, Indonesia) kits. Samples that were positive for Omicron were further investigated by SNP-probe examination, with the target S-gene at amino acid positions 371 and 417. We used a LightCycler 480 II (Roche-Germany) with the following program: activation of the reverse transcriptase at 55°C for 10 minutes; one deactivation cycle at 95°C for 2 min; PCR: one denaturation cycle at 95°C for 10 seconds; annealing at 55°C and reading of the dye signal for one minute, PCR program 45 cycles. Each examination was conducted in triplicate and the results were confirmed by measurements on three different days. The samples tested had Ct values from COVID-19 RT-PCR examination of between 20 and 33. For confirmation, three samples that were positive for the Omicron variant were examined for the whole genome using Oxford Nanopore Technology (ONT)-UK.
Number of Cases in South Tangerang
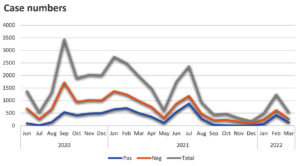
Figure 1 shows the timeline of the COVID-19 cases previously tested from June 2020 to March 2022 in our laboratory. More than 15,000 samples were tested and 6,856 were positive for COVID-19. We obtained samples of COVID-19 patients from hospitals in the region of the Faculty of Medicine, State Islamic University Syarif Hidayatullah Jakarta, namely the South Tangerang area.
Figure 1. The sample numbers of COVID-19 cases in the Laboratory of Microbiology Research, Faculty of Medicine, State Islamic University from June 2020 to March 2022. The grey line is the total number of incoming samples for the PCR test. The orange line denotes the negative results, and the blue line shows the positive results after PCR
Design of Primer and SGTF-SGTP Probe
To detect the SARS-CoV-2 Omicron variant, we designed a primer and specific probe targeting the S-gene. Table 3 shows the amino acid position of SARS-CoV-2 in the S-gene.
Table (3):
The Position of the Amino Acid Changes in the Spike (S) Gene.
| Variant type | Amino acid position in S gene target | |||
|---|---|---|---|---|
| 1 – 300 | 301 – 600 | 601 – 900 | 901 – 1200 | |
| Alpha | Del69-70HV; | N501Y; A570D | D614G; P681H; | S982A; D1118H |
| B.1.1.7 | del144Y | T716I | ||
| Bata | L18F; D80A; | K417N; E484K; | D614G; A701V | – |
| B.1.351 | D215G; del242-244; | N501Y | ||
| R246I | ||||
| Gamma | L18F; T20N; P26S; | K417T; E484K; | D614G; H655Y | T1027I |
| P.1 | D138Y; R190S | N501Y | ||
| Delta | T19R; V70F; T95I; | K417N; L452R; | D614G; H681R | D950N |
| B.1.617.2 | G142D; del156; | T478K | ||
| del157; R158G; | ||||
| A222V; W258L | ||||
| Omicron | A67V;del69/70HV; | G339D; S371L; | D614G; H655Y; | Q954H:N969K |
| B.1.1.529 | T95I; delG142; | S373P; S375F; | N679K; P681H; | |
| delV143; delY144; | K417N; N440K; | N764K; D796Y; | ||
| Y145D; delN211; | G446S; S477N; | N856K | ||
| L212I | T478K; E484A; | |||
| Q493R; G496S; | ||||
| Q498R; T547K | ||||
Note to Table 3: The position of amino acid changes in the spike (S) gene was used as reference for the design of primers and probes
After optimisation, we used the primer and probe to identify the Omicron variant in our samples. We used only samples that had already tested positive for Omicron using commercial kits (SNPsig®-SARS-CoV-2-EscapePLEX and VarScreen-RXReady mBioCoV-19).
Detection of SARS-CoV-2 by Probes 371 and 417 and Confirmed With Established Kit
The 112 samples that tested positive for SARS-CoV-2 in both the RT-PCR test and the variant examination using SNPsig®-SARS-CoV-2 and VarScreen-RXReady were used for the SGTF and SGTP examination of the Omicron variant by SNP-probes with target amino acids S371L and K417N. Figures 2 and 3 show the amplification process for the detection of the Omicron variant using our primers and probes.
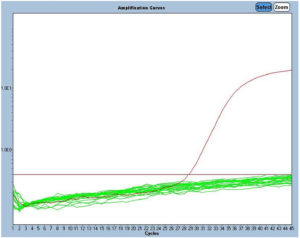
Figure 2. S-gene target failure (SGTF) probe 371; the green lines denote non-amplification; the red line is the amplification graph of g-block Wuhan
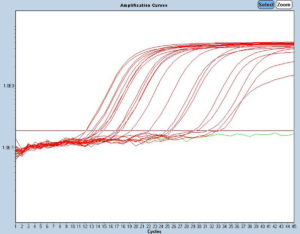
Figure 3. S-gene target positive (SGTP) probe 371; the red lines depict the amplification graphs; the green line denotes non-amplification of g-block Wuhan
Figures 2 and 3 show the SGTF and SGTP examinations. In SGTF, only g-block or synthetic nucleotide Wuhan would amplify with the SGTF probe, whereas the Omicron samples would not. The SGTF probe only binds with the Wuhan nucleotide sequence; if a nucleotide in this sequence is changed, the probe will not bind. No amplification signal will appear if the probe does not bind to the sample. The same principle concerning the probe applies in both SGTP and SGTF; however, the probe will only bind to samples that have the nucleotide sequence of the Omicron variant. Thus, the principle of the SGTP probe is the opposite of that for SGTF.
The selection of target amino acid position 371 was based on the WGS SARS-CoV-2 sequencing of the Omicron variant compared to other variants such as Alpha, Beta, Gamma, My and Delta. In the WGS analysis, the S-gene amino acid target 371 (at the nucleotide position 21,000’s) is in an area of change found exclusively in Omicron and thus not in other variants. As for target amino acid 417, this change is found in Omicron, Delta and Beta.
For the position of the S371L gene, we developed the SGTF and SGTP methods, where SGTF will only be positive for amplification results in the form of Ct and sigmoid curves for non-Omicron variants, while SGTP will only be positive for amplification results in the form of Ct and sigmoid curves for Omicron variants. As for K417N, the method we developed will only detect the Omicron, Delta and Beta variants.
Table (4):
SGTF and SGTP 371/417 confirmation with established kits of SARS-CoV-2 variants.
| No | Sample | SGFT Probe | SGTP Probe | SNPsig EscapePLEX SARS-CoV-2 | VarScreen Rx Ready mBioCoV-19+ | Conclusion | ||
|---|---|---|---|---|---|---|---|---|
| 371L FAM | 417N HEX | 371 FAM | 417N FAM | |||||
| 1 | dH2O | – | – | – | – | – | – | Negative |
| 2 | g-Block Wuhan (Positive) | + | + | – | – | Not test | Not test | COVID-19 Wuhan |
| 3 | g-Block Omi and Delta (Positive) | – | – | + | + | Not test | Not test | COVID-19 Omi/Delta |
| 4 | Positive Control for Prime/Biofarma | Not test | Not test | Not test | Not test | + | + | COVID-19 Omi |
| 5 | 14354 | – | – | + | + | + | + | COVID-19 Omi |
| 6 | 14357 | – | – | + | + | + | + | COVID-19 Omi |
| 7 | 14291 | + | – | – | + | + | + | COVID-19 Delta |
| 8 | 11440 | + | + | – | – | – | – | COVID-19 Wuhan |
| 9 | 10922 | + | + | – | – | – | – | COVID-19 Wuhan |
| 10 | 14280 | + | + | – | – | – | – | COVID-19 Wuhan |
| 11 | 12165 | + | – | – | + | + | + | COVID-19 Delta |
| 12 | 14334 | – | – | + | + | + | + | COVID-19 Omi |
| 13 | 14335 | – | – | + | + | + | + | COVID-19 Omi |
| 14 | 14336 | – | – | + | + | + | + | COVID-19 Omi |
| 15 | 14355 | – | – | + | + | + | + | COVID-19 Omi |
| 16 | 14311 | – | – | + | + | + | + | COVID-19 Omi |
| 17 | 14291 | + | + | – | – | – | – | COVID-19 Wuhan |
| 18 | 14306 | – | – | + | + | + | + | COVID-19 WGS OMI |
| 19 | 14332 | – | – | + | + | + | + | COVID-19 Omi |
| 20 | 14333 | – | – | + | + | + | + | COVID-19 Omi |
| 21 | 14331 | – | – | + | + | + | – | COVID-19 WGS OMI |
| 22 | 14397 | – | – | + | + | + | + | COVID-19 Omi |
| 23 | 14366 | – | – | + | + | + | + | COVID-19 Omi |
| 24 | 14384 | – | – | + | + | + | + | COVID-19 Omi |
| 25 | 14385 | – | – | + | + | + | + | COVID-19 Omi |
| 26 | 14389 | – | – | + | + | + | + | COVID-19 Omi |
| 27 | 14391 | – | – | + | + | + | + | COVID-19 Omi |
| 28 | 14392 | – | – | + | + | + | + | COVID-19 Omi |
| 29 | 14393 | – | – | + | + | + | + | COVID-19 Omi |
| 30 | 14394 | + | + | – | – | – | – | COVID-19 Wuhan |
| 31 | 14395 | + | + | – | – | – | – | COVID-19 Wuhan |
| 32 | 14396 | + | + | – | – | – | – | COVID-19 Wuhan |
| 33 | 14398 | + | + | – | – | – | – | COVID-19 Wuhan |
| 34 | 14399 | + | + | – | – | – | – | COVID-19 Wuhan |
| 35 | 14400 | + | + | – | – | – | – | COVID-19 Wuhan |
| 36 | 14419 | – | – | + | + | + | + | COVID-19 Omi |
| 37 | 14420 | – | – | + | + | + | + | COVID-19 Omi |
| 38 | 14421 | – | – | + | + | + | + | COVID-19 Omi |
| 39 | 14422 | – | – | + | + | + | + | COVID-19 Omi |
| 40 | 14423 | – | – | + | + | + | + | COVID-19 Omi |
| 41 | 14424 | – | – | + | + | + | + | COVID-19 Omi |
| 42 | 14426 | – | – | + | + | + | + | COVID-19 Omi |
| 43 | 14427 | – | – | + | + | + | + | COVID-19 Omi |
| 44 | 14428 | – | – | + | + | + | + | COVID-19 Omi |
| 45 | 14429 | – | – | + | + | + | + | COVID-19 Omi |
| 46 | 14438 | + | + | – | – | – | – | COVID-19 Wuhan |
| 47 | 10061 | + | – | – | + | + | + | COVID-19 Delta |
| 48 | 10922 | + | + | – | – | – | – | COVID-19 Wuhan |
| 49 | 12165 | + | + | – | – | – | – | COVID-19 Wuhan |
| 50 | 13597 | + | – | – | + | + | + | COVID-19 Delta |
| 51 | 13759 | + | + | – | – | – | – | COVID-19 Wuhan |
| 52 | 10058 | + | – | – | + | + | + | COVID-19 Delta |
| 53 | 14764 | – | – | + | + | + | + | COVID-19 Omi |
| 54 | 14931 | – | – | + | + | + | + | COVID-19 Omi |
| 55 | 14820 | – | – | + | + | + | + | COVID-19 Omi |
| 56 | 14747 | + | + | – | – | – | – | COVID-19 Wuhan |
| 57 | 14927 | + | + | – | – | – | – | COVID-19 Wuhan |
| 58 | 14819 | – | – | – | – | – | – | COVID-19 Wuhan |
| 59 | 14680 | – | – | + | + | + | + | COVID-19 Omi |
| 60 | 14697 | – | – | + | + | + | + | COVID-19 Omi |
| 61 | 14699 | – | – | + | + | + | + | COVID-19 Omi |
| 62 | 14712 | – | – | + | + | + | + | COVID-19 Omi |
| 63 | 14744 | – | – | + | + | + | + | COVID-19 Omi |
| 64 | 14747 | – | – | + | + | + | + | COVID-19 Omi |
| 65 | 14748 | – | – | + | + | + | + | COVID-19 Omi |
| 66 | 14759 | – | – | + | + | + | + | COVID-19 Omi |
| 67 | 14764 | – | – | + | + | + | + | COVID-19 Omi |
| 68 | 14779 | – | – | + | + | + | + | COVID-19 Omi |
| 69 | 14780 | – | – | + | + | + | + | COVID-19 Omi |
| 70 | 14781 | – | – | + | + | + | + | COVID-19 Omi |
| 71 | 14782 | – | – | + | + | + | + | COVID-19 Omi |
| 72 | 14783 | – | – | + | + | + | + | COVID-19 Omi |
| 73 | 14729 | – | – | + | + | + | + | COVID-19 Omi |
| 74 | 14638 | – | – | + | + | + | + | COVID-19 Omi |
| 75 | 14656 | – | – | + | + | + | + | COVID-19 Omi |
| 76 | 14661 | – | – | + | + | + | + | COVID-19 Omi |
| 77 | 14662 | – | – | + | + | + | + | COVID-19 Omi |
| 78 | 14663 | – | – | + | + | + | + | COVID-19 Omi |
| 79 | 14664 | – | – | + | + | + | + | COVID-19 Omi |
| 80 | 14665 | – | – | + | + | + | + | COVID-19 Omi |
| 81 | 14666 | – | – | + | + | + | + | COVID-19 Omi |
| 82 | 14677 | – | – | + | + | + | + | COVID-19 Omi |
| 83 | 14678 | – | – | + | + | + | + | COVID-19 Omi |
| 84 | 14681 | – | – | + | + | + | + | COVID-19 Omi |
| 85 | 14682 | – | – | + | + | + | + | COVID-19 Omi |
| 86 | 14683 | – | – | + | + | + | + | COVID-19 Omi |
| 87 | 14594 | – | – | + | + | + | + | COVID-19 Omi |
| 88 | 14498 | – | – | + | + | + | + | COVID-19 Omi |
| 89 | 14526 | – | – | + | + | + | + | COVID-19 Omi |
| 90 | 14527 | – | – | + | + | + | + | COVID-19 Omi |
| 91 | 14552 | – | – | + | + | + | + | COVID-19 Omi |
| 92 | 14553 | – | – | + | + | + | + | COVID-19 Omi |
| 93 | 14509 | – | – | + | + | + | + | COVID-19 Omi |
| 94 | 14510 | – | – | + | + | + | + | COVID-19 Omi |
| 95 | 14511 | – | – | + | + | + | + | COVID-19 Omi |
| 96 | 14512 | – | – | + | + | + | + | COVID-19 Omi |
| 97 | 14513 | – | – | + | + | + | + | COVID-19 Omi |
| 98 | 14514 | – | – | + | + | + | + | COVID-19 Omi |
| 99 | 14515 | – | – | + | + | + | + | COVID-19 Omi |
| 100 | 14486 | – | – | + | + | + | + | COVID-19 Omi |
| 101 | 14496 | – | – | + | + | + | + | COVID-19 Omi |
| 102 | 14497 | – | – | + | + | + | + | COVID-19 Omi |
| 103 | 14542 | – | – | + | + | + | + | COVID-19 Omi |
| 104 | 14529 | – | – | + | + | + | + | COVID-19 Omi |
| 105 | 14554 | – | – | + | + | + | + | COVID-19 Omi |
| 106 | 14555 | – | – | + | + | + | + | COVID-19 Omi |
| 107 | 14556 | – | – | + | + | + | + | COVID-19 Omi |
| 108 | 14557 | – | – | + | + | + | + | COVID-19 Omi |
| 109 | 14478 | – | – | + | + | + | + | COVID-19 Omi |
| 110 | 14487 | – | – | + | + | + | + | COVID-19 Omi |
| 111 | 14596 | – | – | + | + | + | + | COVID-19 Omi |
| 112 | 14542 | – | – | + | + | + | + | COVID-19 Omi |
Table 4 shows the results using our probes and confirmation with the two commercial SARS-CoV-2 variant kits. Here, our method returned similar results to those of the commercial kits and (to the extent it was conducted) the WGS examination of our samples.
Significant findings of our study are : (1). Through the g-block or nucleotide fragment as a positive control, the two types of probes for each target amino acid fit specifically with the ancestral Wuhan or the Omicron variant. These specific results confirmed the correctness and success of the fluorescence signal; (2). Here, our diagnostic method using probe 371 and 417 gave similar results to those of the commercial kits and (to the extent it was conducted) the WGS examination of our samples.
Number of Cases in South Tangerang
Since the first report of SARS-CoV-2 in 2019, there have been many accounts regarding the characteristics of the virus. However, since the virus mutates very easily and changes its properties, there is unlikely to be an end to the discussion around SARS-CoV-2 and its variants.13,14 The most recently reported variant, in late 2021, was Omicron, which can spread rapidly. However, reports stating an increase in severity and mortality in adults due to this mutation compared to the previous Delta variant have been rare.13-20
In December 2021, when cases of the SARS-CoV-2 Omicron variant increased worldwide, the Indonesian Ministry of Health instructed all government referral laboratories to examine this mutation, especially using WGS examination.21,22 However, not all referral laboratories have WGS instruments and sufficient financing for such expensive analysis, so referral laboratories were authorised to test variants using SNP or SGTF kits. Unfortunately, not every RT-PCR machine in the laboratories could be used for variant detection using the commercial test kits provided by the Ministry of Health; the kits could only be used with certain types of RT machines. Therefore, we developed a method for the detection of Omicron variants that can be used in all RT-PCR machines.
In general, the high-cost process of the genome sequencing method is used to trace SARS-CoV-2 variants. Currently, the development of PCR techniques is increasingly rapid and makes it easier to track a nucleotide change in the gene of living organisms. Several conventional PCR methods can be used to track changes in one or more nucleotides, such as restriction fragment length polymorphism (RFLP)-PCR, specific primer-PCR, SNP-PCR and so on, which generally require agarose electrophoresis for detection. These techniques are cheaper but require more time and work, meaning they are very time-consuming if more than 10 samples have to be examined. Other, non-conventional PCR techniques may be suitable, such as RT-PCR, quantitative (q) PCR or digital PCR.23-29
The principle of these techniques is the same as in conventional PCR; to amplify a DNA fragment. In qPCR, the thermocycler has a sensor to detect a fluorescence signal. In addition to a primer, we include a probe, an internal primer with a fluorescence dye or a reagent mix containing the fluorescence solution (such as Sbyr). Thus, we can observe the amplification directly and “real-time in-process” by applying non-conventional PCR methods such as SNP-probe PCR, Rhamp-SNP PCR, Sbyr-PCR and so on.28,29
Design of Primer and SGTF-SGTP Probe
The SGTF and SGTP examination was based on the identification of SNP or double nucleotide polymorphism (DNP) methods.29,30 The position of the nucleotide change can be in the coding, non-coding or intergenic regions.13,28-30 We developed a probe that recognises the amino acid leucine at position 371 in Omicron instead of serine in the ancestral Wuhan variant. As such, the probe will attach to the Omicron variant that has changed from 371 serine to leucine, but there will be no attachment to the ancestral variant or other variants without such substitution. Likewise, the test probe for K417N, which involves the substitution of lysine 417 to asparagine, has the same principle as for S371L, in both SGTF and SGTP.
In our experiments, we designed two types of probes for each target amino acid, namely the ancestral Wuhan and the Omicron variant. With the g-block or nucleotide fragment as a positive control, they fit each type of probe. The existence of two probe models and g-blocks to target the SARS-CoV-2 S-gene further confirmed the correctness and success of the fluorescence signal. For detection, we used carboxyfluorescein (FAM) in a single-plex inspection reaction or a single test. The reason for choosing this fluorescence dye is that FAM sensors can be found in all RT-PCR cyclers. Thus, even in laboratories equipped with old-type RT-PCR instruments and limited signal channels, the method we developed can be considered for detecting Omicron variants.
Detection of SARS-CoV-2 by Probes 371 and 417 and Confirmed With Established Kit
Our SGTF and SGTP results were all confirmed by the same results obtained via tests using SNPsig and VarScreen reagents (Table 4). This indicates that the probes we tested can be applied for trace examinations of SARS-CoV-2 Omicron variants. In addition to using an existing kit, we confirmed the results using Oxford Nano Technology genome sequencing, demonstrating success in probe testing. Therefore, our design probe can be used not only in Indonesia but also globally. The limitation of our study derived from the patient data (hospital records) as we could only obtain swab samples with numbers and no information regarding the condition of the patients. Our laboratory is not a hospital laboratory but a medical faculty research laboratory that is seconded for COVID-19 examination in the South Tangerang area. Thus, we do not hold patient data. It would have been preferable for the hospital that sent the sample to have also provided complete data on the patient’s status.
In conclusion, our research successfully developed an SNP identification method for SARS-CoV-2 variants using specific probes for SGTF/SGTP S371L and K417N. Our method can be used for the detection of SARS-CoV-2 Omicron variants.
ACKNOWLEDGMENTS
The authors would like to thank the members of the COVID-19 laboratory Faculty of Medicine Syarif Hidayatullah State Islamic University Jakarta, Ministry of Health Republic Indonesia, PT. Biopharma and PT. Elokarsa Utama for their support in conducting the research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
CA conceptualized the study, applied methodology and performed validation. CA, EAS, LAH, ZH, S, DFR, AL, FE, EW, FRS, SK, MAAF and DRN performed analysis. CA and HJF wrote the original draft. HJF, EAS, LAH, ZH, S, DFR, AL, FE, EW, FRS, Sk, MAAF and DRN reviewed and revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Ethics Committee, Faculty of Medicine UIN Syarif Hidayatullah Jakarta (No. B-005/F12/ KPK/TL.00/02/2021).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924.
Crossref - COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. https://github.com/CSSEGISandData/COVID-19. Accessed June 11, 2022
- Indonesia National Board for Disaster Management: https://covid19.go.id/peta-sebaran. Accessed June 11, 2022
- Harvey WT, Carabelli AM, Jackson B, et al. COVID-19 Genomics UK (COG-UK) Consortium. SARS-CoV-2 variants, spike mutations, and immune escape. Nat Rev Microbiol 2021;19(7):409-424.
Crossref - Otto SP, Day T, Arino J, et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol. 2021;31(14):R918-R929.
Crossref - Muhawarman A. Waspada Varian Baru Omicron. 2021. https://mediakom.kemkes.go.id/2021/12/waspada-varian-baru-omicron/ Accessed June 11, 2022
- Kwok PY, Chen X. Detection of Single Nucleotide Polymorphisms. Curr Issues Mol Biol. 2003;5(2):43-60. PMID:12793528. https://pubmed.ncbi.nlm.nih.gov/12793528/ Accessed June 11, 2022
- Wang E, Adams S, Zhao Y, et al. Methodology: A strategy for detection of known and unknown SNP using a minimum number of oligonucleotides applicable in the clinical settings. J Transl Med. 2003;1(1):4.
Crossref - Tabit FT. Advantages and limitations of potential methods for the analysis of bacteria in milk: a review. J Food Sci Technol. 2016;53(1):42-49.
Crossref - Gupta PK, Roy JK, Prasad M. Single nucleotide polymorphisms: A new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Current Science. 2001;80(4):524-35. https://www.jstor.org/stable/24104242 Accessed June 11, 2022
- Gonzalez-Bosquet J, Chanock SJ. Basic principles and laboratory of genetic variation. IARC Sci Publ. 2011;163: 99-120. PMID:22997858. Accessed June 11, 2022
- Kadri K. Polymerase Chain Reaction (PCR): Principle and Applications. Synthetic Biology – New Interdisciplinary Science. IntechOpen. 2019.
Crossref - Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450-452.
Crossref - Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity, and global spread of SARS-CoV-2. Nature. 2021;600(7889):408-418.
Crossref - Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7889):21.
Crossref - Nyberg T, Ferguson NM, Nash SG, et al. COVID-19 Genomics UK (COG-UK) consortium. Comparative analysis of the risks of hospitalization and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303-1312.
Crossref - Kumar S, Thambiraja TS, Karuppanan, Subramaniam G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of the spike protein. J Med Virol. 2022;94(4):1641-1649.
Crossref - Sigal A, Milo R, Jassat W. Estimating disease severity of Omicron and Delta SARS-CoV-2 infections. Nat Rev Immunol. 2022;22(5):267-269.
Crossref - European Centre for Disease Prevention and Control. SARS-CoV-2 variants of concern as of 12 May 2022. https://www.ecdc.europa.eu/en/covid-19/variants-concern Accessed June 11, 2022
- European Centre for Disease Prevention and Control. Methods for the detection and characterization of SARS-CoV-2 variants – first update. https://www.ecdc.europa.eu/en/publications-data/methods-detection-and-characterisation-sars-cov-2-variants-first-update Accessed June 11, 2022
- CDC Covid-19 Response Team. SARS-CoV-2 B1.1.529 (Omicron) variant – the United States, December 1-8, 2021. Morb Mortal Wkly Rep. 2021;70(50):1731-1734.
Crossref - Hemarajata P. SARS-CoV-2 Sequencing Data: The Devil is in the Genomic Detail. American Society Micro. 2020. https://asm.org/Articles/2020/October/SARS-CoV-2-Sequencing-Data-The-Devil-Is-in-the-Gen Accessed June 11, 2022
- Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34(3):275-305.
Crossref - Jehan T, Lakhanpaul S. Single nucleotide polymorphism (SNP) – methods and applications in plant genetics: A review. Indian J Biotech. 2006;5(4):435-459. http://nopr.niscpr.res.in/handle/123456789/5608 Accessed June 11, 2022
- Mackay IM, Arden KE, Nitsche A. Real-time Fluorescent PCR Techniques to Study Microbial – Host Interactions. Methods Microbiol. 2004;34:255-330.
Crossref - Adhiyanto C, Hendarmin L, Puspitaningrum R, Hendarto H. Pengenalan Dasar Teknik Bio-Moleculer (Introducing Basics of Bio-Molecular Techniques). Yogyakarta, Deepublish;c2019:23-59.
- Bhagwat M. Searching NCBI’s dbSNP database. Curr Protoc Bioinformatics. 2010;Chapter 1(Unit-1.19).
Crossref - Brown KA, Gubbay J, Hopkins J, et al. S-Gene Target Failure as a Marker of Variant B.1.1.7 Among SARS-CoV-2 Isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA. 2021;325(20):2115-2116.
Crossref - Rosenfeld JA, Malhotra AK, Lencz T. Novel multi-nucleotide polymorphisms in the human genome characterized by whole genome and exome sequencing. Nucleic Acid Res. 2010;38(18):6102-6111.
Crossref - Zou H, Wu LX, Tan L, Shang FF, Zhou HH. Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs. Front Cell Dev Biol. 2020;8:347-361.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.