ISSN: 0973-7510
E-ISSN: 2581-690X
The present work was conducted to isolate and detect by PCR three important bacterial pathogens (Staphylococcus aureus, Streptococcus agalactiae, and Streptococcus uberis) associated with bovine mastitis. A total of 36 clinical mastitic milk samples were collected from various cross bred cattle presented in the Veterinary Clinical complex centre, Bikaner, Rajasthan. Out of 36 milk samples processed, we recovered 51 isolates belonging to Staphylococcus aureus (18), Streptococcus uberis (5), Streptococcus agalactiae (2), Staphylococcus intermedius (8), Staphylococcus chromogens (4), Bacillus sp. (2) and Escherichia coli (12). The PCR conducted directly on milk samples with species-specific primers of Staphylococcus aureus, Streptococcus agalactiae and Streptococcus uberis revealed 19, 7 and 2 samples respectively positive for these pathogens. The study showed highest incidence of Staphylococcus aureus (50.0%) followed by Streptococcus uberis, (13.89%) and Streptococcus agalactiae (5.5%) by culture-based method while incidence rate recorded by PCR of Staphylococcus aureus (52.77%) followed by Streptococcus uberis, (19.44%) and Streptococcus agalactiae (5.5%) associated with bovine mastitis. Thus, PCR was found to have a good correlation with cultural method for diagnosis of the causative agent of mastitis in a short span of time.
Mastitis, cattle, PCR, Staphylococcus, Streptococcus
Mastitis is one of the important challenges to the dairy industry in both developed and developing countries like India. It affects the economy of the farmers and the country, accounting for an annual estimated loss of around US$526 million (Dua, 2010). The noteworthy increase in the incidence of bovine mastitis is an alarming phase for the dairy sector. More than 200 infectious causes of mastitis are known to date. The commonest pathogens in large animals are Staphylococcus aureus, Streptococcus agalactiae, Streptococcus uberis and Coliforms in Asia (Chahar et al., 2008; Yong et al., 2009). To treat, monitor and control mastitis, there is a great need to identify the etiological agents rapidly and accurately.
Conventional in vitro culture and biochemical tests used are time-consuming, laborious and not highly specific. Moreover, sub-clinically infected animals intermittently shed the bacteria that may yield no bacteria with milk culture (Phuektes et al. 2001). To overcome the limitations of cultural and biochemical identification of organisms, the molecular methods of identification of bacteria based on amplification of specific segments of the genome of bacteria by polymerase chain reaction have been developed and found useful as these are more reliable and less laborious (Anand-Kumar, 2009; Kozytska et al. 2010). Thus the present study was started to investigate the prevalence of various important bacterial agents associated with bovine mastitis and use PCR for their rapid and accurate detection.
The milk samples were collected from various cross-bred cattle presented in the from Veterinary Clinical Complex, College of Veterinary and Animal Science, Bikaner showing symptoms of hard and swelled udder, change in the appearance of the milk and presence of clots. The teats were cleaned with a cotton swab soaked in 70% alcohol. The first few streams of mastitic milk were discarded and then 2 – 4 ml of secretion was collected in sterile test tubes. Milk samples were immediately taken thereafter to the laboratory on ice for direct DNA extraction and bacterial culturing. The DNA was extracted directly from milk using Phenol chloroform method with isopropanol precipitation (Murphy et al., 2002). Also, milk samples were inoculated on a variety of medias viz Blood Agar, Mannitol Salt agar and MacConkeys Agar for isolation and identification of bacterial species as per the standard protocol (Quinn and Carter, 1994).
The DNA extracted directly from milk was subjected to Polymerase chain reaction for detection of three main mastitis causing bacterial species viz. S. aureus, S. agalactiae, and S. uberis using already published primers of Phuektes et al. (2001) which are given in Table 1.
Table (1):
Primer sequences, annealing temperatures and size of the amplified products of various primers used in this study.
Species |
Sequence (5′-3′) |
Annealing temperature (°C) |
Product size (bp) |
|---|---|---|---|
S. aureus |
TCT TCA GAA GAT GCG GAA TA TAA GTCAAACGTTAACATACG |
56 |
420 |
S. agalactiae |
AAG GAAACCTGCCATTTG TTA ACCTAGTTTCTTTAAAACTAGAA |
55 |
270 |
S. uberis |
TAA GGAACACGTTGGTTAAG TTCCAGTCCTTAGACCTTCT |
55 |
330 |
The PCR reaction was carried out in 25 µl reaction volume in 0.2 ml PCR tubes using the Promega gene amplification kit (Madison, Wisconsin, USA) by mixing 6 µl 5X assay buffer, 1 µl F-primer (10 pM/µl), 1 µl R-primer (10 pM/µl), 1 µl dNTP (10 mM), 3 µl MgCl2 (1.5 mM/µl), 0.25 µl Taq DNA polymerase (5 U/µl), 14.75 µl deionised water, and 3 µl template DNA (25 ng/µl). The PCR was performed in Eppendorf Mastercycler Gradient (Eppendorf AG, Hamburg, Germany). The amplification reaction for S. aureus involved initial denaturation at 96°C for 1 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, and final extension of 72°C for 10 min. Reaction conditions and PCR master mix concentrations were similar for Streptococcus agalactiae and S. uberis except the annealing temperature was 55°C. The PCR products were analyzed by electrophoresis on 2% agarose gel run along with 250 bp DNA ladder (Promega, USA) and amplicons were visualized under UVP Gel Doc Bioimaging System (UVP LLC, Upland, CA).
In the present investigation out of the total 36 milk samples processed a total of 51 isolates belonging to four different genera were recovered. Out of 51 isolates, 25 isolates were found positive for targeted bacterial species viz. Staphylococcus aureus and Streptococcus sp whereas rest of the samples revealed the growth of Escherichia coli. Out of 25 samples which were positive for targeted bacterial species, 14 samples showed presence of mix cultures of Staphylococcus sp., Streptococcus sp. and Bacillus sp. while 7 samples showed pure growth of S. aureus, 2 for S. agalactiae and 2 for S. uberis. From 14 mix culture revealing samples, it was found 5 samples contained S. aureus in addition to S. intermedius, 2 samples contained cultures of S. aureus along with Bacillus sp., 4 were having S. aureus along with S. chromogens and 3 samples revealed the presence of S. uberis with S. intermedius. The growth of Staphylococcus aureus on Mannitol salt agar showed typical yellow discolouration of the media indicating mannitol fermenting capacity of the isolates which is indirectly related to pathogenicity of the strains. Streptococcus sp. showed pin-point, dew-drop like colonies on Blood agar and revealed the typical Gram-positive bacteria in short chains.
The PCR conducted directly on milk samples with species-specific primers of Staphylococcus aureus, Streptococcus agalactiae and Streptococcus uberis revealed 19, 7 and 2 samples respectively positive for these pathogens (Fig. 1).
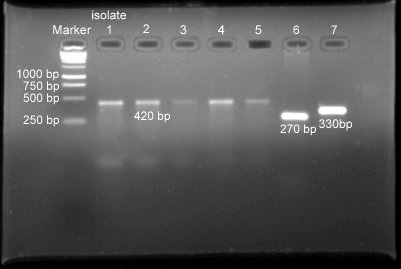
Fig. 1. Lane 1- Ladder, Lane 2-5- S. aureus (420bp), Lane 6 – S. agalactiae (270bp), Lane7- S. uberis (330bp)
The study found that all those samples which were positive by cultural method were also positive by PCR for Staphylococcus aureus, Streptococcus agalactiae, and Streptococcus uberis. Thus, the PCR method can be successfully applied for speedy diagnosis of the mastitis eliminating the need of lengthy protocols for isolation and biochemical confirmation. However, as the PCR is not able to differentiate between dead and viable bacteria, the cultural method cannot be replaced entirely. As in the present study, there was one milk sample which was found positive for S. aureus by PCR did not reveal any growth of S. aureus. Similarly, two milk samples were positive by PCR for S. uberis which could not show any growth of this bacterium. This can be attributed to dead/non-viable bacteria present in the milk sample which could not be revived during isolation. However, the PCR is based on the presence of specific template DNA which can be either from dead or viable bacteria.
Mastitis is the most common infectious disease affecting dairy cows (Birthal and Jha, 2005) and remains a major challenge to worldwide dairy industry despite the widespread implementation of mastitis control strategies (Oviedo-Boyso et al., 2007). Staphylocococcus aureus has been the most important bacteria responsible for various forms of mastitis globally including India. Several authors have recorded the high prevalence of the bacterium from different parts of the world which are in well accordance with our findings (Mekkib et al., 2010; Ranjan et al., 2011). Streptococci are the second group of microorganisms in importance, after Staphylococci, responsible for mastitis in ruminants (Bergonier et al. 1999). Streptococcus agalactiae, S. uberis, and S. dysgalactiae are the species frequently identified in mammary gland infections. In the present study we recovered only two species of Streptococcus with a prevalence of 13.89% of S. uberis and 5.5% of S. agalactiae. S. uberis which was recovered in high number in the present study, correlates with the report of Denis et al. 2009 who found it to be most abundant streptococcal species from mastittic milk in New Zealand. The prevalence rate of S. uberis was very much close to the findings of Bradley (2002) who had reported a prevalence of 12.7% of the bacterium. S. agalactiae was the second important streptococcal species recovered from mastitic milk samples which have been reported with similar isolation rate by Khaled et al. (2010). The study showed a high recovery of E. coli (36.1%) which can be attributed to poor sanitation conditions prevailing in the sheds and of milkers leading to environmental mastitis (Bradley, 2002).
To overcome the limitations of cultural and biochemical identification of organisms, the molecular methods of identification of bacteria based on amplification of specific segments of the genome of bacteria by PCR have been developed. We found the high accuracy of the PCR in rapid detection of some important bacterial pathogens. This helped in reducing the considerable work load which is done in accurate detection of bacterial species by various cultural, primary and secondary biochemical tests. The present investigation finds the great applicability of PCR in solving the problem of identification within a short span of time.
- Anand-Kumar, P. Evaluation of PCR test for detecting major pathogens of Bubaline mastitis directly from mastitic milk samples of buffaloes. Trop Ani Health Prod, 2009: 41: 1643-1651.
- Bergonier, D.X., Berthelot, M., Romeo, A., Contreras, V., Coni, E., De Santis, S., Roselu, F., Barillet, G., Lagriffoul, Marco, J. Fréquence des différents germes responsables de mammites cliniques et subcliniques chez les petits ruminants laitiers, In F. Barillet and P. Zervas (ed.), Milking and milk production of dairy sheep and goats. 1999, Wageningen Pers, Wageningen, The Netherlands. pp. 130–136.
- Birtlial, P.S., Jha, A.K. Economic losses due to various constraints in dairy production in India. Indian Journal of Animal Sciences, 2005, 75(12): 1470-1475.
- Bradley, A.J. Bovine mastitis an evolving disease. The Veterinary Journal, 2002: 164: 116-128.
- Chahar, A., Gahlot, A.K., Tanwar, R.K., Fakhruddin. Evaluation of different screening tests for diagnosis of sub clinical mastitis in cattle. Indian J. Vet. Med, 2008: 28: 91-93.
- Denis, M., Wedlock, D.N., Lacy-Hulbert, S.J., Hillerton, J.E., Buddle, B.M. Vaccines against bovine mastitis in the New Zealand context: What is the best way forward? New Zealand Veterinary Journal, 2009: 57: 132–140.
- Dua, K. Incidence, etiology and estimated economic losses due to mastitis in Punjab and India. An update. Indian Dairyman, 2010: 53: 41–48.
- Khaled, A., El-Razik, A., Abdurrehman, K.A., Ahmed, Y.F., Gomaac, A.M. Eldebaky, HA. Direct Identification of Major Pathogens of the Bubaline Subclinical Mastitis in Egypt using PCR. Journal of American Science, 2010: 6: 652-660.
- Kozytska, S., Staub, D., Pawlik, M.C., Hensen, S., Eckart, M., Ziebuhr, W., Witte, W., Ohlsen, K. Identification of specific genes in Staphylococcus aureus strains associated with bovine mastitis. Vet Microbiol, 2010: 145: 360-365.
- Mekibib, B., Furgasa, M., Abunna, F., Megersa, B., Regassa, A. Bovine Mastitis: Prevalence, Risk Factors and Major Pathogens in Dairy Farms of Holeta Town, Central Ethiopia. Veterinary World, 2010: 3(9): 397-403.
- Murphy, M. A., Shariflou, M. R., Moran, C. High quality genomic DNA extraction from large milk samples. Journal of Dairy Research, 2002: 69: 645–649.
- Oviedo-Boyso, J., Valdez-Alarcon, J.J., Cajero-Juarez, M., Ochoa-Zarzosa, A., López-Meza, J.E., Bravo-Patino, A., Baizabal-Aguirre, V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. Journal of Infection, 2007: 54(4): 399-409.
- Phuektes, P., Mansell, P.D., Browning, G.F. Multiplex polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and Streptococcal causes of bovine mastitis. Journal of Dairy Science, 2001: 84: 1140-48.
- Quinn, P.J., Carter, M.E., Markey, B.K., Carter, G.R. Clinical Veterinary Microbiology, London:Wolfe Publishing, Mosby-Year Book Europe Ltd. 1994, pp 7-12.
- Ranjan, R., Gupta, M.K., Singh, K.K. Study of bovine mastitis in different climatic conditions in Jharkhand, India. Veterinary World, 2011: 4(5): 205-208.
- Sun, S., Negrea, A., Rhen, M., Andersson, D.Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother, 2009: 53(6): 2298-305.
- Yong, Z., Fang, J.X., Mei, Y., Narisu, S., Zhong, L.B. Isolation and identification of pathogens from mastitis cow and drug sensitivity test. China Anim. Husb. Vet. Med, 2009: 36: 136-140
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


