ISSN: 0973-7510
E-ISSN: 2581-690X
Shiga toxin-producing Escherichia coli (STEC) can cause from watery diarrhea to severe diseases, such as thrombocytopenic purpura (TP) or hemolytic uremic syndrome (HUS). Ruminants are the main reservoir; most of the time humans are infected through the ingestion of contaminated meat. The goal of this study was the detection of stx1 and stx2 genes through the standardization of a real time PCR method that uses SYBR green, and then we used this method to analyze the presence of STEC in ground beef from butcheries of the metropolitan zone of Asuncion, Paraguay. We analyzed 48 ground beef samples from 24 butcheries. They were processed by standard methods of culture and DNA extraction. Real-time PCR reactions were standardized for the detection of stx1 and stx2. Both genes were detected in the 48 samples studied. The concordance between this method and a conventional PCR technique was excellent, showing kappa concordance indexes of 0.76 for stx1 and 0.87 for stx2. Even though the high levels of contaminated ground beef detected, all the isolates were LEE negatives and O157 negatives, which are less related to outbreaks and severe diseases. The most common virulence profile was stx1/stx2/saa/exhA. With this finding, we have proven the need for preventive procedures in the slaughter process and the handling process of meat products.
STEC; ground beef; LEE-negative STEC.
Shiga toxin-producing E. coli (STEC), also known as verotoxigenic E. coli (VTEC) and enterohemorrhagic E. coli (EHEC), is one of the most important groups of food borne pathogens1,2. Its main characteristic is that it carries the genes stx1 and stx2, these genes codify cytotoxins, which are considered essential virulence factors that cause the pathologies in humans3,4.
It can cause watery diarrhea, bloody diarrhea, thrombocytopenic purpura, hemor-rhagic colitis (CH), and hemolytic uremic syndrome (HUS)5,6. There are different combinations of genetic markers between different STEC strains, but it is still unclear which one is necessary to produce pathologies7. However, the food safety organizations classify as high risk those strains that carry the following virulence genes: stx/eae, and stx/aggR/aaiC. High-risk strains are more likely to cause outbreaks and severe diseases8,9.
While STEC O157-H7 is the most common serotype reported in outbreaks and severe disease cases10,11; currently other non-O157 serogroups (O26, O45, O103, O111, O145, and O104) are also recognized as important causes of human pathologies3,12,13.
Ruminants are asymptomatic reservoirs of these pathogens, so meat products and their derivatives represent the main contamination sources in outbreaks and cases of human infection14,15.
Meat production is an axis of Paraguayan economy. It has occupied the first places in the exportation rankings. Also, it is placed third with the highest consumption of meat worldwide (25,6kg per citizen), according to the latest report of the Organization for Economic Cooperation and Development (OECD)17.
The difficulties in controlling the meat expenditure are the high amount and diversity of butcheries for the local population, as well as the differences in regulations between the several agencies. All these facts highlight the need for the implementation of methods that allow the rapid and sensitive detection of meat contaminated with STEC strains. Real-time polymerase chain reaction (qPCR) is an excellent option to solve this need. For all of the above, the goal of this study was the detection of stx1 and stx2 genes through the standardization of a real-time PCR method that use SYBR green, and then we used this method to analyze the presence of STEC in ground beef from butcheries of the metropolitan zone of Asuncion, Paraguay.
Controls
We used as positive controls previously characterized E. coli isolates with differences virulence genes, STEC no-O157 carries stx1 and stx2, STEC O157: H7 carries stx2 and O157, STEC O157: H7 carries stx1 and O157 as positive controls, they were kindly donated by Dr. Noralia Padola, Universidad Nacional del Centro de la Provincia de Buenos Aires.
Butcheries selection
Butcheries were selected after the implementation of an audit system that evaluated risk factors in three main components of the shops: building, equipment, and instruments, as well as workers hygienic habits. To this end, we used a validated tool that was previously implemented in Argentina, under the name of ‘’Carnicerias saludables’’ (‘’Healthy butcheries’’), and it has been adapted for this study17.
This tool allowed us to classify butcheries as high, moderate or low risk of selling products contaminated with pathogens. In this study, we included 24 butcheries that were classified as high risk of selling contaminated products.
Sample collection
250 g of ground beef were collected twice in each of the butcheries; the second collection was three months later than the first. The samples were transported refrigerated to the laboratory, where they were coded and processed.
Bacteriological procedure and Sample processing
Ten grams of ground beef in 100 mL of buffered peptonated water were incubated overnight at 37°C. Two aliquots were taken for DNA extraction and storage. Non-contaminated samples of ground beef were injected with STEC strains, to be able to use them as positive extraction controls. The artificially contaminated meat was processed in the same way as the samples. Additionally, negative controls were added to each trial.
DNA extraction
DNA extraction was performed following the manufacturer’s instructions using a commercial kit (Wizard Genomic, Promega, USA).
DNA quantification
It was performed by absorbance measure at 260 nm and 280 nm using a DNA quantifier (Biowave DNA, WPA, UK).
Sample storage
Bacterial culture was cryopreserved at -80°C in vials with BHI medium (Brain Heart Infusion) (Britania, Argentina) with 15% glycerol (Lasca, Paraguay). DNA aliquots from each sample were cryopreserved at -20°C.
Real-time PCR standardization
Oligonucleotides and reaction conditions described by Brusa et al were used. They reported a limit of detection of 1×102 CFU / mL, as well as inclusivity and exclusivity of 100%7.
Melting temperature (Tm) of PCR products was determined experimentally using as positive controls three strains that carriers stx1, stx2, and stx1 y stx2 genes respectively, this temperature was corroborated theoretically using OligoCalc (program). Samples were considered positive when they generated the same melting curve as the positive control. Results from real-time PCR were compared with the results from a multiplex PCR previously described7.
Bacterial isolates, serogroups and virulence factors detection
Three positive samples with the shorter cqs were selected and grew in Luria Bertani (LB) agar plates at 37°C for 24 hours. 20-50 isolates were analyzed from selected samples. DNA was extracted from each isolate that was previously grown in liquid LB at 37°C for 24 hours; virulence factors genes (stx1,stx2, eae, exhA, saa); and serogroups(O26, O45, O103, O104, O111, O121, O145 y O157, O78) were detected, following the conditions previously described2,18–21.
Real-time PCR standardization
A sigmoidal amplification curve was observed for both genes in ground beef that was artificially injected with STEC strains, and no amplification was observed in the negative control. The melting curve showed a single peak at 79°C in concordance with the theoretical value. Minor peaks were not observed, these generally are associated with the formation of primer dimers or other secondary products (Fig. 1).
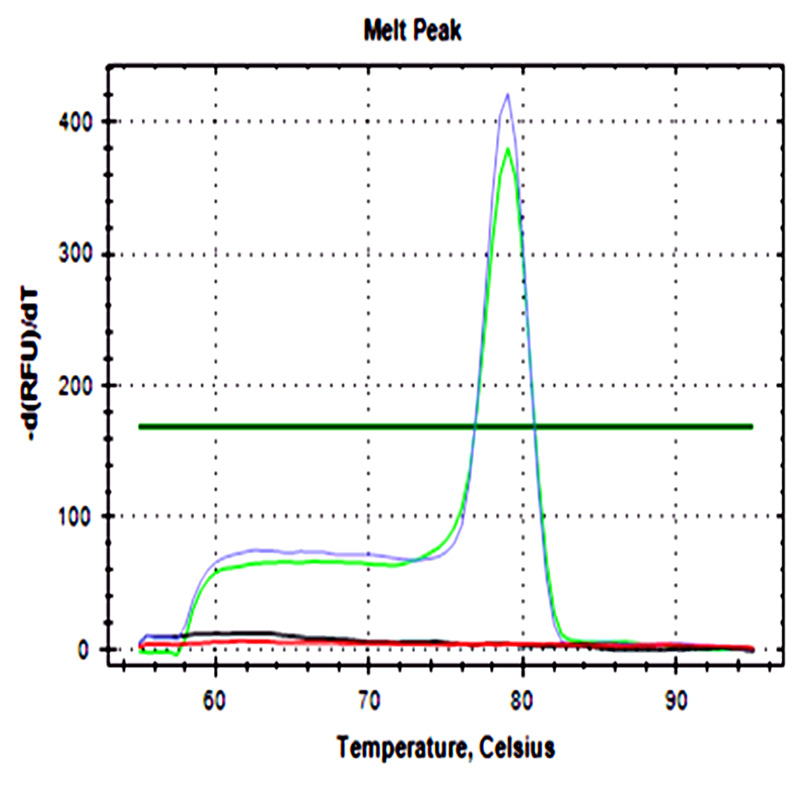
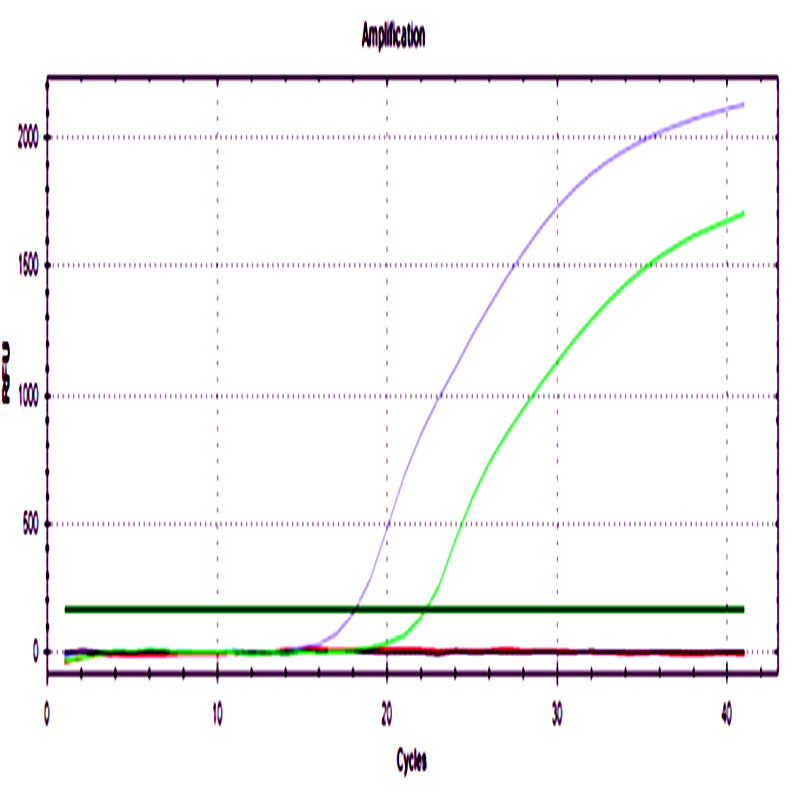

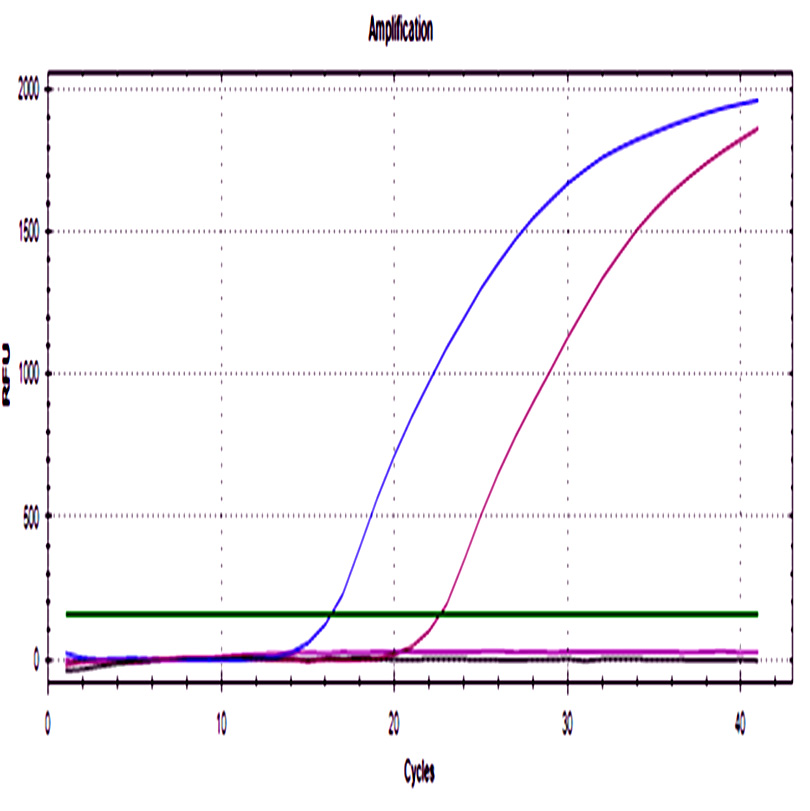
Fig. 1. a) Amplification curve of the stx1 b) The melting curve of the stx1 gene. c) Amplification curve of the stx2 gene. d) Melting curve of the stx2 gene. Blue curves represented STEC NOO157 (stx1/stx2), green and purple curves represent meat injected with positive control, black curves represent negative controls of PCR assay, and red curves represent negative controls of the extraction process.
Cross-reactions test
To verify that the technique did not give false positives through cross-reactions, we included four STEC strains as controls with the following genetic profiles stx1, stx2, stx1/stx2, and an E. coli strain without stx genes respectively. No cross-amplifications were observed between stx1 and stx2 genes.
stx1/stx2 detection in ground beef
The 48 ground beef samples showed fluorescence peaks with the same melting curve of positive controls. No amplification was detected in negative controls from ADN extraction and real-time PCR assay.
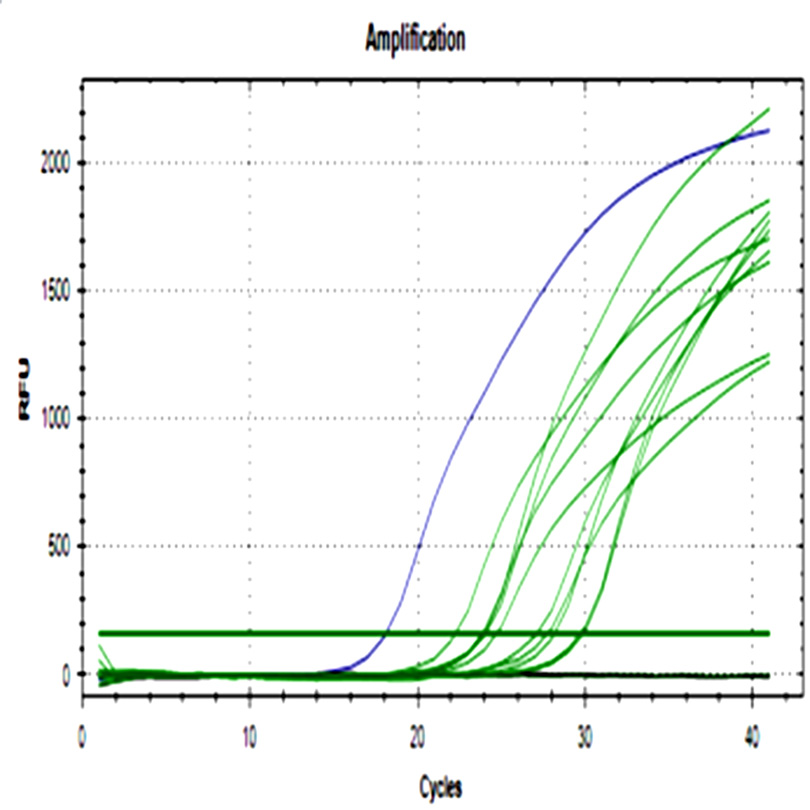
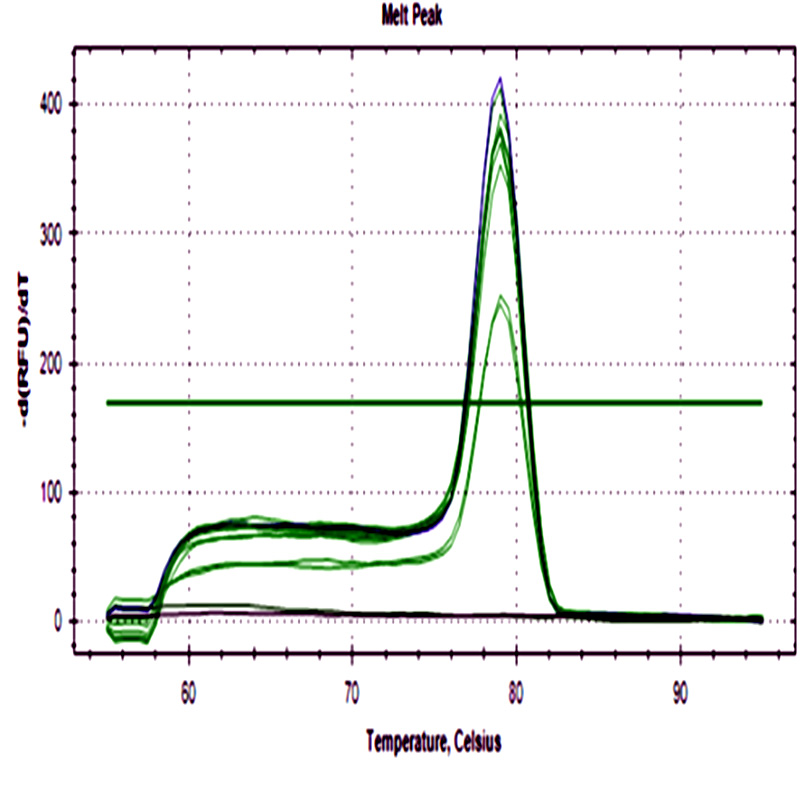
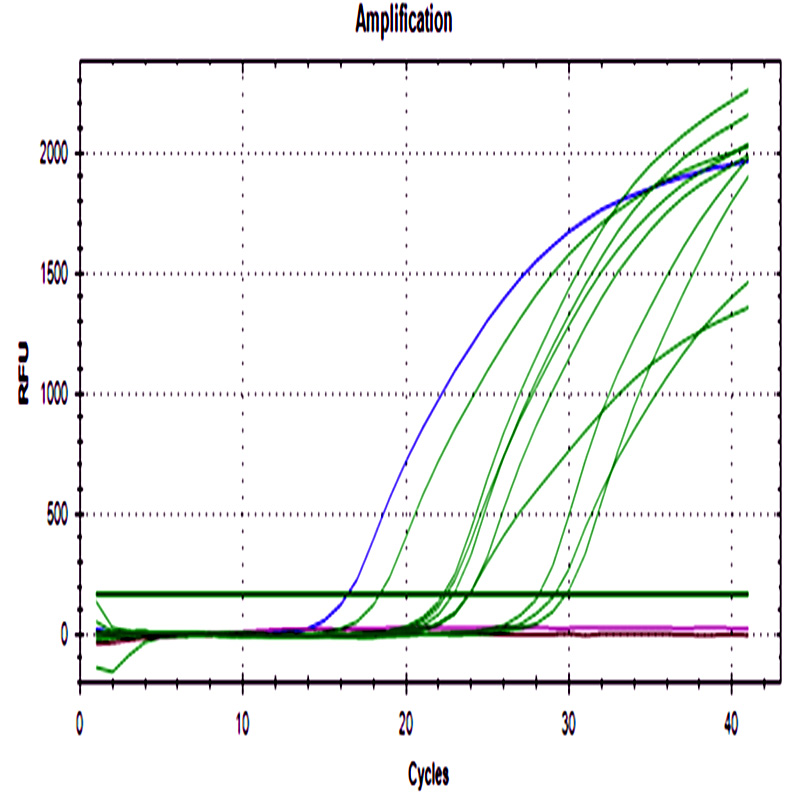
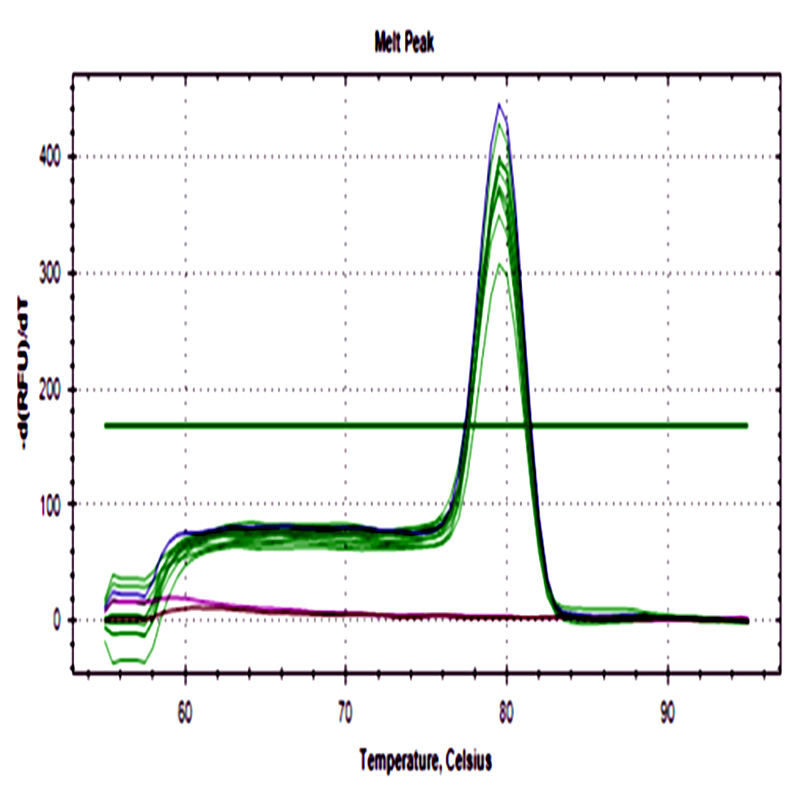
Fig. 2. a) Amplification curve of stx1 gene. b) The melting curve of stx1. c) Amplification curve of stx2. d) Melting curve of stx2. Blue curves represent positive control, red curves represent negative control of PCR assay, black curves represent negative control of the DNA extraction process, and green curves ground beef samples from different butcheries.
All samples were positive for both genes with a wide cq range between 14.6- 34.4 for stx1, and 15- 32.2 for stx2 demonstrating differences in the initial bacteria amount among them.
stx1/ stx2 amplification was performed by a multiplex PCR in the same ground beef samples; this technique was previously developed and used in the laboratory. An excellent concordance between real-time PCR and multiplex PCR results was observed with a kappa index of 0.76 for stx1. For this gene, four discordances were recorded, four positive samples for real-time PCR were negative samples for Multiplex PCR, all of them with prolonged cq values of 29.8 to 32.6. For the stx2 gene, the highest concordance between the methods was observed with a kappa index of 0. 86, with two in consistencies that were positive samples for real-time PCR and negative ones for multiplex PCR. The cq of the discordant samples were also prolonged 29.8 and 29.9 respectively.
rfbO157 detection in ground beef
rfbO157 was not detected by a conventional PCR in any sample.
STEC isolates
Real-time PCR results were confirmed by isolation of STEC strains from selected ground beef samples then their genetic characterization was done by conventional PCR. Three positive samples from different butcheries were selected because they presented the lowest values of cq. The analysis of a total of 150 isolates allowed the detection of 12 STEC strains, any of them have the eae gene which is why they were classified as LEE-negative (without the locus of enterocyte effacement), also O157-negative, serogroups O26, O45, O103, O104, O111, O121, O145 were not detected either. The virulence profiles were stx1/stx2/saa/exhA in 9 isolates, stx2/saa/exhA in two isolates and stx1/saa/exhA in one isolate. Real-time PCR results were confirmed by a multiplex PCR that detects other regions of the stx1 and stx2 genes, as well as standard microbiological methods for identification of viable STEC isolates, which virulence profiles were detected by molecular methods.
Our previous evaluation of butcheries showed shortcomings in infrastructure, as well as failures in good hygiene practices at the local shops. The fact that all butcheries were classified as high risk of selling products that could be contaminated with pathogens17, highlights the need for control systems based on the education of sellers and consumers about standard operating methods. These results were consistent with the findings of STEC in all ground beef samples.
Real-time PCR for STEC detection showed advantages like a sensitive and specific amplification of target genes without interferences of the complex matrix. This fact has been demonstrated by the successful detection of target genes in artificially contaminated meat. Ground beef is a very complex sample because it has a lot of amplification inhibitors, very low bacterial load, and high background of other bacteria, so finding the right extraction method is complicated22.
Most of the commercial real-time PCR kits are based on probes labeled with fluorophores, allowing simultaneous detection of different targets9, being much more expensive than those based on DNA intercalary agents. This fact could be an important factor for the selection of screening method due to a large number of samples to be analyzed and the low-resource in a developing country. Previous studies showed comparable results between real-time PCR techniques that based on probes and those with SYBR Green9,11,22,23, Brusa et al. found a correlation between RT-PCT and SYBR-PCR of 0.758 in stx1 and 0.802 in stx2(p<0.01)22.
Real-time PCR results showed a high concordance with multiplex PCR results for both genes. In fact, it showed that the real-time PCR has a higher sensitivity than the conventional PCR, being able to detect both genes in samples that were negative for multiplex PCR, due to a very low amount of gene targets revealed by cq values between 29 and 359,11,14,15,24,25. This high sensitivity represents an advantage of this technique due to pathogenic STEC having a very low infectious dose, 10 to 100 bacteria per gram of food could produce disease1,10,14. At this moment control systems need to apply sensitive screening methods to evaluate meat contamination.
In this study, the rfbO157 gene was not detected in any sample. This serotype was barely reported in Paraguay, limited to specific reports of 4% of E. coli O157: H7 in food, and a clinical case of STEC O157: H7 in a child with HUS26,27. Even though STEC O157 was not detected; this doesn’t mean that there is no risk of outbreaks when contaminated meat products are on sale.
The quality criteria demanded by international control agencies have changed due to the emergence of hybrid strains such as STEC/EAEC strain that caused one of the biggest outbreaks registered in Europe in 201113. Those new criteria have included in the analysis of meat products the detection of other serotypes in addition to the O157: H7, as well as virulence genes profile of strains[7].There are more than 400 non-O157 serotypes that caused human diseases and outbreaks. They were found in different reservoirs such as water, meat, and vegetables, thus environments, which are in frequent contact with humans15.
It’s essential to isolate STEC strains in samples that were positive for stx1 andstx2 by molecular methods, to confirm if pathogens are viable in the sample, and also to know their virulence profile as it provides a lot of information about its pathogenicity3,7. However, bacterial isolation from meat samples is a challenge for a lot of factors such as scarce phenotypic differentiation between E. coli and other species such as Shigella spp, low initial STEC load, and high background of other species that inhibit the growth of the target3,7,14,28.
Microbiological analysis of positive ground beef samples with the lowest ct values showed bacteria viability and allowed isolation of 12 STEC non-O157 strains, virulence gene profiles that included combinations of stx1/stx2/saa/ehxA, stx1/saa/ehxA, and stx2/saa /ehxA. None of these isolates belonged to higher clinical relevant serogroups O26, O45, O103, O104, O111, O121, O145,and O157; neither have eae, so they were classified as LEE-negative. These findings highlight that we need to apply standard operative practices and strengthen our control systems to avoid the contamination of meat products. Even with these troubling results, it is important to emphasize that EFSA classified LEE-negative strains as low risk of causing outbreaks and severe infection. This is positive for the meat industry, although it remains necessary to take preventive measures mainly in animal slaughtering process and meat product handling.
Our previous evaluation results base on risk factors in building, equipment, and instruments, as well as workers hygienic habits of butcheries had a high concordance with molecular analysis of ground beef samples. This evaluation protocol represents an accessible and cheap tool that could be used to audit butcheries, applied by control systems as well as owners for internal control. We proposed to use this simple tool to classify the risk levels of butcheries, to be able to work on improving places with high risk and workers education.
None
The authors declare no conflict of interest.
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Escherichia coli. Nat. Rev.2004; 2: 123–140, doi:10.1038/nrmicro818.
- Parma, A.A.E.; Sanz, M.E.; Blanco, J.E.; Blanco, J.; Viסas, M.R.; Blanco, M.; Echeverría, A.I.; European, S.; Parmal, A.E.; Sanz, M.E.; et al. Virulence Genotypes and Serotypes of Verotoxigenic Escherichia coli Isolated from Cattle and Foods in Argentina/ : Importance in Public Health Published by/ : Springer Stable URL/ : http://www.jstor.org/stable/3582458 Accessed/ : 13-07-2016 23/ : 01 UTC Virule. Eur. J. Epidemiol., 2000; 16: 757–762.
- Brusa, V.; Pinero, P.E.; Galli, L.; Linares, L.H.; Ortega, E.E.; Padola, N.L.; Leotta, G.A. Isolation of Shiga Toxin-Producing Escherichia coli from Ground Beef Using Multiple Combinations of Enrichment Broths and Selective Agars. Foodborne Pathog. Dis., 2016; 13: 163–170, doi:10.1089/fpd.2015.2034.
- Fratamico, P.M.; Bagi, L.K. Detection of Shiga toxin-producing Escherichia coli in ground beef using the GeneDisc real-time PCR system Bacterial Strains And Growth Conditions. Front. Cell. Infect. Microbiol., 2012; 2: 1–6, doi:10.3389/fcimb.2012.00152.
- Karmali, M.A. Emerging Public Health Challenges of Shiga Toxin-Producing Escherichia coli (STEC) related to Changes in the Pathogen , the Population , and the Environment. Clin. Infect. Dis., 2016; 1–22.
- Angelika, A.; Rita, F.; Erhard, P.; Wolfgang, T.; Flieger, R.A. Molecular epidemiological view on Shiga toxin-producing Escherichia coli causing human disease in Germany: diversity, prevalence, and outbreaks. Int. J. Med. Microbiol., 2015; 5: 1–26, doi:10.1016/j.ijmm.2015.08.020.
- Brusa, V.; Galli, L.; Linares, L.H.; Ortega, E.E.; Linn, J.P.; Leotta, G.A. Development and validation of two SYBR green PCR assays and a multiplex real-time PCR for the detection of Shiga toxin-producing Escherichia coli in meat. J. Microbiol. Methods, 2015; 119: 10–17, doi:10.1016/j.mimet.2015.09.013.
- BIOHAZ Scientific Opinion on VTEC-seropathotype and scientific criteria regarding. EFSA J., 2013; 11: 3138, doi:10. 2903/j.efsa.2013.3138.Available.
- Kagkli, D.-M.; Folloni, S.; Barbau-Piednoir, E.; Van den Eede, G.; Van den Bulcke, M. Towards a Pathogenic Escherichia coli Detection Platform Using Multiplex SYBR®Green Real-Time PCR Methods and High Resolution Melting Analysis. PLoS One, 2012; 7: e39287, doi:10.1371/journal.pone. 0039287.
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev., 1998; 11: 142–201, doi:file://Z:\References\Text Files\00000004472.txt.
- Kagkli, D.; Weber, T.P.; Bulcke, M. Van Den; Folloni, S.; Tozzoli, R.; Morabito, S.; Ermolli, M.; Gribaldo, L.; Eede, G. Van Den Application of the Modular Approach to an In-House Validation Study of Real-Time PCR Methods for the Detection and Serogroup Determination of Verocyto-toxigenic Escherichia coli. Appl. Environ. Microbiol., 2011; 77: 6954–6963, doi:10.1128/AEM.05357-11.
- Ferreira, M.R.A.; Filho, E.G.F.; Pinto, J.F.N.; Dias, M.; Moreira, C.N. Isolation, prevalence, and risk factors for infection by shiga toxin-producing Escherichia coli (STEC) in dairy cattle. Trop. Anim. Health Prod., 2014: 46, 635–639, doi:10.1007/s11250-014-0541-5.
- Dakshina M. Jandhyala, PhD, Vijay Vanguri, MD, Erik J. Boll, PhD, YuShuan Laic, B.; A. McCormick, PhD, and John M. Leong, MD, P. Shiga Toxin–Producing Escherichia coli O104:H4: An emerging pathogen with enhanced virulence. Infect Dis Clin North Am., 2014; 27: 631–649, doi:10.1016/j.idc.2013. 05.002.Shiga.
- Linares, L.H.; Sanz, M.E.; Echeverría, A.I.; Padola, N.L.; Galli, L.; García, P.P.; Copes, J.; Leotta, G.A. Shiga toxin-producing Escherichia coli in beef retail markets from Argentina. Front. Cell. Infect. Microbiol., 2013; 2: 1–6, doi:10.3389/fcimb.2012.00171.
- Padola, N.L. Advances in detection methods for Shiga toxin-producing Escherichia coli (STEC). Front. Microbiol., 2014; 5: 1–2, doi:10.1086/338115.
- Litvinoff, N. Consumo interno de carne crecerá un 5,7% este ano. La Naciףn 2016, http://www.lanacion.com.py/2016/09/07/consumo-inte.
- Vet, M.; Ortega, E. Carnicerías saludables Dirigido a autoridades bromatológicas; 2014.
- Blanco, M.; Blanco, J.E.; Mora, A.; Dahbi, G.; Alonso, M.P.; Gonza, E.A.; Blanco, J. Serotypes , Virulence Genes , and Intimin Types of Shiga Toxin (Verotoxin) -Producing Escherichia coli Isolates from Cattle in Spain and Identification of a New Intimin Variant Gene (eae- xi). J. Clin. Microbiol., 2004; 42: 645–651, doi:10.1128/JCM.42.2.645.
- Padola, N.L.; Sanz, M.E.; Blanco, J.E.; Blanco, M.; Blanco, J.; Etcheverria, I.; Arroyo, G.H.; Usera, M.A.; Parma, A.E. Serotypes and virulence genes of bovine Shigatoxigenic Escherichia coli (STEC) isolated from a feedlot in Argentina. Vet. Microbiol., 2004; 100: 3–9, doi:10.1016/S0378-1135(03)00127-5.
- Rumi, M.V.; Irino, K.; Deza, N.; Huguet, M.J.; Bentancor, A.B. First isolation in Argentina of a highly virulent Shiga toxin-producing Escherichia coli O145/ : NM from a domestic cat. J Infect Dev Ctries, 2012; 6: 358–363.
- Sanchez, S.; Llorente, M.T.; Echeita, M.A.; Herrera-Lean, S. Development of three multiplex PCR assays targeting the 21 most clinically relevant serogroups associated with Shiga toxin-producing E. coli infection in humans. PLoS One, 2015; 10: 1–11, doi:10.1371/journal.pone.0117660.
- Brusa, V.; Linares, L.H.; Ortega, E.E.; Lir, J.P.; Leotta, G.A. Development and validation of two SYBR green PCR assays and a multiplex real-time PCR for the detection of Shiga toxin-producing Escherichia coli in meat. J. Microbiol. Methods, 2015; doi:10.1016/j.mimet.2015.09.013.
- Friedrich, A.W.; Borell, J.; Bielaszewska, M.; Fruth, A. Shiga Toxin 1c-Producing Escherichia coli Strains/ : Phenotypic and Genetic Characterization and Association with Human Disease. J. Clin. Microbiol., 2003; 41: 2448–2453, doi:10.1128/JCM.41.6.2448.
- Yves Germani, Patrick Cunin, Etienne Tedjouka, Choua bou Ncharre, Jacques Morvan, Paul Martin. Escherichia coli in Ngoïla ( Cameroon ) during an outbreak of bloody diarrhea. The Lancet 1998; 352.
- Tanaro, J.D.; Leotta, G.A.; Lound, L.H.; Deza, N.; Ledri, S.E.; Carbonari, C.; Piaggio, M.C. Deteccion de Escherichia coli productor de toxina Shiga a partir de bovinos, agua ambiental y muestras clinicas (Gualeguaychú, Entre ríos). Ciencia, Docencia, y Tecnol. Supl., 2013; 3: 1–17.
- Copes, J.; Pellicer, K.; Hoyo, G.; M, L.C.; Estigarribia, M.; G, P.O.; Loup, V.; Florentin, C.; Alonso, M.; Cardozo, L. Primer aislamiento de Escherichia coli O157/ : H7 a partir de hamburguesas en Paraguay First Isolation of Escherichia coli O157/ : H7 from hamburgers in Paraguay. Analecta Vet., 2009; 29: 11–14.
- Noceda, L. Síndrome urémico hemolítico. Pediatr. (Asuncion), 2009; 36: 131–137.
- Jure, M.A.; Condori, S.; Leotta, G.A.; Chinen, I.; Miliwebsky, E.; Allori, C. productor de toxina Shiga a partir de carne molida fresca proveniente de carniceras de Concepción, provincia de Tucuman. Rev. Argent. Microbiol., 2010; 2010: 284–287.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


