ISSN: 0973-7510
E-ISSN: 2581-690X
Mycoplasma pneumoniae is recognized as the leading cause of community-acquired lower respiratory tract infection in children, accounting for a significant proportion of pediatric mortality. Macrolides are the first-line treatment for M. pneumoniae infections. However, the extensive use of macrolides in clinical practice resulted in the emergence of macrolide-resistant M. pneumoniae (MRMP), which has a negative impact on treatment outcomes. Hence, in the present study, MRMP was determined in hospitalized children with community-acquired pneumonia (CAP). Real-time PCR detected M. pneumoniae in 41 of 348 clinical samples. Sanger sequencing revealed that none of the isolates were associated with the A2063G or A2064G base mutation, which confers macrolide resistance, in domain V of the 23S rRNA gene. Although MRMP was not observed in children with CAP in our study, healthcare practitioners should be vigilant about the potential risk of MRMP infections.
Macrolide Resistant Mycoplasma pneumoniae, Community-acquired Pneumonia, 23S rRNA Gene Sequencing
Community-acquired pneumonia (CAP) is the leading cause of hospitalizations, with substantial morbidity and a high mortality rate in children.1 Bacterial pathogens were considered one of the major causes of CAP, with Mycoplasma pneumoniae responsible for 10–40% of CAP infections in children worldwide.2,3 The CAP caused by M. pneumoniae was underestimated due to its mild symptoms, being overshadowed by other bacterial illnesses, and having limited rapid diagnostic methods. The World Health Organization (WHO) estimates M. pneumoniae is responsible for 20% of childhood mortality.4 In Southeast Asia, M. pneumoniae is responsible for 6.8% and 22% of deaths in neonates and children aged under 5 years, respectively.5 Most cases of childhood pneumonia in India are caused by bacteria; nevertheless, M. pneumoniae is one of the major respiratory pathogens causing atypical pneumonia in children.4 Waning immunity is believed to be a major cause of recurring infections in children, and it is therefore endemic in nature irrespective of risk factors. M. pneumoniae infection can be effectively managed with early detection and adequate antibiotic therapy. M. pneumoniae infection can be effectively managed with early detection and adequate antibiotic therapy. The beta-lactam group of antibiotics was ineffective against the cell wall, M. pneumoniae. Protein synthesis inhibitors, such as macrolides, are recommended as first-line antibiotics for M. pneumoniae infection. The overuse of macrolide antibiotics has led to the emergence of macrolide resistant M. pneumoniae (MRMP) strains caused by mutations in domain V of 23S rRNA, resulting in varying degrees of macrolide resistance. The prevalence of these MRMP infections has steadily increased during the last decade. It varies between 80 and 95 percent, especially in the Asian subcontinent.6 Rates have escalated to more than 90% in China, Japan, and Korea,6 more than 30% in Israel, and 12–13% in the United States and Canada.7 Despite the fact that the MRMP is increasing in other parts of the world, limited data is available in our region. Regular surveillance of M. pneumoniae associated with CAP will provide data on the impact of infection on children. Hence, in the present study, the prevalence of MRMP infection was determined among the hospitalised children with pneumonia in our region.
Patient population
The prospective study was conducted from September 2019 to July 2021. A total of 348 hospitalized children, aged between 2 months to 12 years, at a tertiary care centre in Chennai, South India, were included in the study. Children with the symptoms of community-acquired pneumonia as per the WHO criteria were included in the study (WHO, 2014). Children with aspiration pneumonia, hospital-acquired pneumonia, congenital heart diseases, airway anomalies, cystic fibrosis, congenital and acquired immunodeficiency were excluded from the study. The institutional human ethical committee approved our study, and informed consent was obtained from the children’s parents before collecting the samples.
Sample collection and DNA isolation
Sputum samples were collected from children diagnosed with CAP and transported immediately to the laboratory at 4°C. Further, DNA was extracted from the sputum sample using the QIAamp DNA mini kit (Qiagen, Germany) as per manufacturer instructions. The extracted DNA samples were stored at -20°C until further processing.
Mycoplasma pneumoniae detection by RT-PCR
RT-PCR assays, which include primers (5′-GTGAACGTATCGTAACACGAGCTTT-3′ and 5′-TGGTTTGTTGACTGCCACTGC-3′) and probes [5′FAMTTGTCGCGCACTAAGGCCCACG-BHQ-1-3′] that were specific to a 125-bp fragment of the P1-cytadhesin gene, were used to confirm the presence of M. pneumoniae in the clinical samples. The reaction was performed on a QuantStudio3 Flex real-time PCR machine (Thermo Fisher Scientific, Inc.) using a total reaction volume of 10 μL containing probe fast universal master mix(KAPA. REF: KK4702), 10 picomoles each of forward and reverse primers, and a 10ng DNA template. For RT-PCR, the following thermocycling conditions were used: 3 minutes at 95˚C, 3 seconds at 95˚C followed by 20 seconds at 60˚C and 30 seconds at 60˚C for 40 cycles.
Screening of macrolide resistant Mycoplasma pneumoniae
MRMP was determined by amplifying and sequencing the domain V region of the 23S rRNA gene using the primers and PCR conditions described in previous literature.8 The Big Dye Terminator cycle sequencing kit (version 3.1; catalogue number 4337454; Applied Biosystems Inc.) was used to perform the sequencing reaction on the sense strand of the amplicon under the following thermal conditions: 1 cycle at 96°C for 1 min, followed by 25 cycles at 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. The PCR products were purified by ethanol precipitation, and the ABI 3130XL instrument (Applied Biosystems Inc.) was used to sequence them under conditions appropriate for a 50-cm capillary. The raw sequencing data was analysed using the Bioedit programme to identify the gene mutations. Appropriate positive and negative controls were included in the study.
Statistical Analysis
Descriptive statistics were employed to analyse and summarize the important features of the study data.
Detection of M. pneumoniae
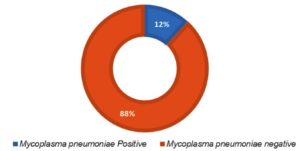
M. pneumoniae was observed in 11.7% (41/348) children presented with CAP using real-time PCR. Figure 1 shows the prevalence of M. pneumoniae.
Clinical manifestations of children infected with M. pneumoniae
The majority of the M. pneumoniae positive samples (53.66%) were observed among the male children. Children aged one to five years have a higher prevalence of children with M. pneumoniae infection (63.41%) compared to other age groups were represented in the Table 1. Simultaneously, Table 2 shows the clinical symptoms such as cold, cough, breathlessness, chest in drawing and fever were observed among the children tested positive for M. pneumoniae infection. The results of X-rays showed that 58.6% of children had lobar pneumonia and bronchopneumonia, and that 17 out of 41 children (41.4%) who had a negative chest X-ray for pneumonia had tested positive for M. pneumoniae in a real-time PCR experiment. This emphasises the superiority of molecular detection over other traditional methods.
Table (1):
Table representing the proportions of age and sex of the M. pneumoniae positive and negative samples.
| Prevalence of Mycoplasma pneumoniae | ||
|---|---|---|
| Mycoplasma infection | ||
| Age & sex | Positive (%) N=41 |
Negative (%) N=307 |
| Gender | ||
| Male | 22 (53.66) | 169 (55.05) |
| Female | 19 (46.34) | 138 (44.95) |
| Age | ||
| < 1 year | 2 (4.87) | 32 (10.43) |
| 1-5 years | 26 (63.41) | 165 (53.74) |
| > 5 years (5-12) | 13(31.71) | 110 (35.83) |
Table (2):
Table representing the proportions of clinical parameters of the M. pneumoniae positive and negative samples.
| Mycoplasma infection | ||
|---|---|---|
| Clinical Parameter | Positive (%) N=41 |
Negative (%) N=307 |
| Cough & Cold | ||
| Present | 35 (85.37) | 231 (75.24) |
| Nil (Absent) | 6 (14.63) | 76 (24.76) |
| Fever duration | ||
| Present | 39 (95.12) | 296 (96.42) |
| Nil (Absent) | 2 (4.88) | 11 (3.58) |
| Breathlessness | ||
| Present | 30 (73.17) | 193 (62.87) |
| Nil (Absent) | 11 (26.83) | 114 (37.13) |
| Chest indrawing | ||
| Present | 10 (24.39) | 97 (31.6) |
| Nil (Absent) | 31 (75.61) | 210 (68.4) |
Mycoplasma pneumoniae 23S rRNA domain V gene sequence analysis by Sanger sequencing
PCR amplification and sequencing of the domain V region of the 23S rRNA gene were analyzed for A2063G and A2064G mutations. Figure 2 represents a multiple sequence alignment file of the domain V region of the M. pneumoniae 23S rRNA gene. In our investigation, there are no variations and all strains are of the wild type.
The current study developed an internally regulated real-time PCR test that targets the P1 adhesion gene to diagnose M. pneumoniae infections. Fluorescent probes improve PCR specificity without requiring post-PCR processing and significantly reduce laboratory result turn around time, which improves patient management. In the present study, M. pneumoniae infection was observed in 11.7% of the study population, which is consistent with the previous study.9 Macrolides are widely used first-line medications for the treatment of M. pneumoniae, which is responsible for 40% of CAP worldwide and 30% of CAP in India.10-12
Our investigation revealed that M. pneumoniae infection primarily occurs in children aged between 1-5 years, which was consistent with the previous reports.13-15 Previous studies showed a significant difference in gender-based approaches, were girls being more susceptible to M. pneumoniae infection than boys.16 On the contrary, we found that male children had a higher incidence than female children, which could be attributed to differences in ethnicity and lifestyle.
In addition to molecular detection, the study examined the clinical subjects’ chest X-rays and found that among the M. pneumoniae positive cases, 43.9% of children had lobar pneumonia compared to other pneumonia, which is consistent with the findings of previous study that found a positive correlation between the aetiology of lobar pneumonia and M. pneumoniae infection.17
The study also compared the association of clinical symptoms with M. pneumoniae infection, but we did not find any notable change due to the similarity in signs and symptoms displayed by other typical and atypical pathogens. This misinterpretation will lead to antibiotic mismanagement. Thus, advancements in diagnostic techniques are required for specific detection of M. pneumoniae, which will undoubtedly aid in antibiotic selection.18,19
Macrolides are widely used as first-line medications for M. pneumoniae infection which accounts for 40% of CAP worldwide and 30% of CAP in India.10-12 Although tetracyclines, fluoroquinolones and erythromycin are effective in treating M. pneumoniae infection, they are not recommended for use in children due to their toxicity. Until the rise of the drug resistance in 2000, macrolides were the potential antibiotic in treating pneumonia caused by M. pneumoniae.2,20 The first MRMP strain was observed in Japan, and since then, the use of macrolides has begun to be reduced as a result of increased resistance, which has become a global burden.2,20,21. The majority of MRMP strains have 23S rRNA gene mutations (2063G and A2064G), which have been identified as one of the major mechanisms by which M pneumoniae acquires macrolide resistance.22,23 In the present study, no mutations in domain V of 23S rRNA gene were observed in M. pneumoniae, indicating that MRMP is not present in our study population. This suggests that we are in the early stages of macrolide resistance, which may worsen over the next decade. Regardless of the situation in India, the rapid spread of MRMP strains throughout Asia is a major concern. To reduce antibiotic misuse, continuous epidemiological monitoring of macrolide resistance must be prioritised.24. As far as we know, this is the first study in our region to perform direct screening for MRMP in sputum of children with CAP. In the current study, induced sputum from children was found to be more representative of the lower respiratory tract than nasopharyngeal aspirates, which may later indicate the carrier state.
Real-time PCR is a sensitive, specific and rapid method for diagnosing M. pneumoniae infection in children. Although macrolide resistance was not observed in our study, given the unscrupulous use of macrolides in LRTIs, it may be the beginning of the development of macrolide resistance, which will likely increase over the next decade. Rapid detection of resistant-associated mutations would aid in prescription of an alternate antibiotic regimen, particularly in cases of chronic or recurring M. pneumoniae infection. Furthermore, macrolide stewardship is required to limit the use of these antibiotics and avoid excessive antibiotic prescribing, particularly in countries where macrolide resistance rates are still low. Even though the mutations A2063G and A2064G were not observed in our study, other mutations in the 23S rRNA are still needed to confirm the absence of MRMP in our region.
ACKNOWLEDGMENTS
The authors would like to thank all the parents of the children agreeing to provide clinical samples for this study. We would also like to extend our sincere gratitude to DHR-MRU, University of Madras, for supporting us with the infrastructural facility.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Human Ethics Committee, Dr. ALM PG IBMS, University of Madras, Taramani Campus, Chennai, India, with approval number UM/IHEC/FRM/2021-X.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Zhu YG, Tang XD, Lu YT, Zhang J, Qu JM. Contemporary Situation of Community-acquired Pneumonia in China: A Systematic Review. J Transl Int Med. 2018;6(1):26-31.
Crossref - Lee JK, Lee JH, Lee H, et al. Clonal Expansion of Macrolide-Resistant Sequence Type 3 Mycoplasma pneumoniae, South Korea. Emerg Infect Dis. 2018;24(8):1465-1471.
Crossref - Lanata MM, Wang H, Everhart K, Moore-Clingenpeel M, Ramilo O, Leber A. Macrolide-Resistant Mycoplasma pneumoniae Infections in Children, Ohio, USA. Emerg Infect Dis. 2021;27(6):1588-1597.
Crossref - Yadav RK, Kumar D, Singh A, Ziauddin M, Singh DK. Clinical and microbial spectrum of community-acquired pneumonia in children of north India. Trop Doc. 2021;51(1):71-77.
Crossref - Sonawane AA, Shastri J, Bavdekar SB. Respiratory Pathogens in Infants Diagnosed with Acute Lower Respiratory Tract Infection in a Tertiary Care Hospital of Western India Using Multiplex Real Time PCR. Indian J Pediatr. 2019;86(5):433-438.
Crossref - Chen YC, Hsu WY, Chang TH. Macrolide-Resistant Mycoplasma pneumoniae Infections in Pediatric Community-Acquired Pneumonia. Emerg Infect Dis. 2020;26(7):1382-1391.
Crossref - Hung HM, Chuang CH, Chen YY, et al. Clonal spread of macrolide-resistant Mycoplasma pneumoniae sequence type-3 and type-17 with recombination on non-P1 adhesin among children in Taiwan. Clin Microbiol Infect. 2021;27(8):1169.E1-1169.E6.
Crossref - Wolff BJ, Thacker WL, Schwartz SB, Winchell JM. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob Agents Chemother. 2008;52(10):3542-3549.
Crossref - Wu HM, Wong KS, Huang YC, et al. Macrolide-resistant Mycoplasma pneumoniae in children in Taiwan. J Infect Chemother. 2013;19(4):782-786.
Crossref - Qu J, Chen S, Bao F, Gu L, Cao B. Molecular characterization and analysis of Mycoplasma pneumoniae among patients of all ages with community-acquired pneumonia during an epidemic in China. Int J Infect Dis. 2019;83:26-31.
Crossref - Kumar S, Bharti PK, Baveja CP, Mantan M, Saigal SR, Garg IB. Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of Mycoplasma pneumoniae in pediatric community-acquired lower respiratory tract infections. Indian J Med Microbiol. 2022;40(2):250-253.
Crossref - Pooja PB, Tejashree A, Narayanappa D. Seroprevalence of Mycoplasma pneumoniae and Clinical Profile of Affected Patients in a Tertiary Care Hospital. J Clin Diagn Res. 2019;13(4):DC01-DC04.
Crossref - Chen A, Song L, Chen Z, et al. Immunoglobulin M profile of viral and atypical pathogens among children with community acquired lower respiratory tract infections in Luzhou, China. BMC Pediatr. 2019;19(1):280.
Crossref - Sondergaard MJ, Friis MB, Hansen DS, Jorgensen IM. Clinical manifestations in infants and children with Mycoplasma pneumoniae infection. PloS One. 2018;13(4):e0195288.
Crossref - Kutty PK, Jain S, Taylor TH, et al. Mycoplasma pneumoniae Among Children Hospitalized With Community-acquired Pneumonia. Clin Infect Dis. 2019;68(1):5-12.
Crossref - Zhang Y, Huang Y, Ai T, Luo J, Liu H. Effect of COVID-19 on childhood Mycoplasma pneumoniae infection in Chengdu, China. BMC Pediatr. 2021;21(1):202.
Crossref - Miyashita N, Sugiu T, Kawai Y, et al. Radiographic features of Mycoplasma pneumoniae pneumonia: differential diagnosis and performance timing. BMC Med Imaging. 2009;9:7.
Crossref - Cheong KN, Chiu SS, Chan BW, To KK, Chan EL, Ho PL. Severe macrolide-resistant Mycoplasma pneumoniae pneumonia associated with macrolide failure. J Microbiol Immunol Infect. 2016;49(1):127-130.
Crossref - Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin Microbiol Rev. 2017;30(3):747-809.
Crossref - Lee H, Choi YY, Sohn YJ, et al. Clinical Efficacy of Doxycycline for Treatment of Macrolide-Resistant Mycoplasma pneumoniae Pneumonia in Children. Antibiotics. 2021;10(2):192.
Crossref - Pereyre S, Goret J, Bebear C. Mycoplasma pneumoniae: Current Knowledge on Macrolide Resistance and Treatment. Front Microbiol. 2016;7:974.
Crossref - Cao B, Zhao CJ, Yin YD, et al. High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin Infect Dis. 2010;51(2):189-194.
Crossref - Nummi M, Mannonen L, Puolakkainen M. Development of a multiplex real-time PCR assay for detection of Mycoplasma pneumoniae, Chlamydia pneumoniae and mutations associated with macrolide resistance in Mycoplasma pneumoniae from respiratory clinical specimens. SpringerPlus. 2015;4:684.
Crossref - Gullsby K, Bondeson K. No detection of macrolide-resistant Mycoplasma pneumoniae from Swedish patients, 1996-2013. Infect Ecol Epidemiol. 2016;6:31374.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.