ISSN: 0973-7510
E-ISSN: 2581-690X
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) has become the most dangerous viral infection worldwide. Since its identification in late 2019, the number of medical trials to combat the infection has sharply increased. Here, we investigated the profiles of IgG and IgM in 85 patients with confirmed SARS-CoV-2 infection from day 1 after symptom onset until day 35 with 5-day intervals. Serum samples were collected and stored until use. We observed that IgM levels were detectable on day 5 post symptom onset and increased sharply, with the highest rate detected in moderate cases (32.332 ± 4.32, n=10). Subsequently, a significant reduction in IgM was observed until it was undetectable on day 35 after symptom onset. Meanwhile, IgG levels were detected on day 10 post symptom onset, and the highest rate was observed in moderate cases (8.232 ± 2.3, n=10). A significant increase in IgG rate was observed in all patients, with the highest rate in moderate cases (42.432 ± 4.34, n=67) on day 35 post symptom onset. The statistical difference between the case and control groups was significant (p≤0.001). Two out of 85 patients died during the study.
SARS-COV-2, COVID-19, Serologic response, IgG, IgM
A new outbreak of coronavirus (severe acute respiratory syndrome coronavirus 2, SARS-COV-2) that causes severe pneumonia started in late 2019.1,2 All age groups are vulnerable to SARS-COV-2, causing infectious diseases from mild to moderate and severe cases.3,4 SARS-COV-2 causes the coronavirus disease 2019 (COVID-19) that has been declared a pandemic due to the spread of COVID-19 cases worldwide, with an increasing number of fatalities. Viral infections have been declared as a pandemic when the cases spread all over the world with an increasing number of COVID-19 fatalities. COVID-19 was observed to be more contagious than those with severe acute respiratory syndrome (SARS-COV) and Middle East respiratory syndrome.5,6 COVID-19 is diagnosed using real-time polymerase chain reaction (RT-qPCR) amplification of the viral RNA and computerized tomography (CT) scans of the lungs with transparent lesions.7,8 The sensitivity of these diagnostic tools are relatively high. However, diagnosis is time-consuming and expensive, and a low viral load might also give a false negative result for SARS-COV-2.8-10 Therefore, a rapid serological detection of IgG and IgM specific to viral spike glycoprotein and nucleocapsid is introduced.11,12 It relies on the detection of the viral nucleocapsid (N) and spike glycoprotein (S). Antibody profiling in patients with COVID-19 might be beneficial to understand the antibody response against SARS-COV-2 and the virus-host interaction in patients tested negative in RT-PCR and those with asymptomatic infections.7,12,13 This method is used worldwide to diagnose patients with COVID-19 especially those who tested negative using reverse transcription techniques. However, cross-reactivity of SARS-COV-2 antibody with antibodies against other pathogens, such as SARS-COV and other seasonal coronaviruses, is possible.14-16 Furthermore, the serological detection of anti-SARS-COV-2 IgG and IgM is still unclear and poorly understood.17
The production of humoral immunity components, including IgG and IgM, protects the body from viral invasion. However, excessive response might damage the tissues; for instance, IgG response increases lung inflammation.18,19 Moreover, hyperinflammatory response might affect several organs, such as the kidney and liver, and cause organ failure that may result in heart failure and death.20,21
In this study, we screened the IgG and IgM profiles in patients with COVID-19. A total of 85 patients infected with SARS-COV-2 visited hospitals presenting with mild to severe symptoms. Some of the patients were hospitalized in urgent care, and some needed mechanical ventilation. Serum samples were collected from day 1 of viral infection and upon symptom onset until recovery.
Study design
A total of 85 patients with COVID-19 confirmed using reverse transcription polymerase chain reaction (RT-PCR) volunteered to participate in this study. All cases were confirmed to be infected using routine clinical testing, such as monitoring of symptoms, RT-PCR, and CT scan of the patient’s chest. The study was conducted between September 15, 2020 and February 1, 2021. All patients were admitted to specialty hospitals in Mosul City, Iraq. The age of the patients was between 35 and 72 years (average age, 45 years). Patients were observed to have moderate to severe symptoms and critical symptoms requiring respiratory mechanical ventilation. The patients were grouped according to the following criteria: moderate cases are defined as having high fever, fatigue symptoms, and pneumonia on lung radiography; severe cases should show respiratory distress saturation with aerial partial pressure symptoms; and critical cases should show respiratory failure requiring mechanical ventilation and exhibit multiple organ dysfunction symptoms. Serum samples were collected from all patients from day 1 of symptoms onset until recovery with 5 day intervals, having a total of eight serum samples from each patient. Ten serum samples from healthy individuals were included as controls. All serum samples were stored at −19°C until further use.
Antibody detection
Anti-spike-glycoprotein (S) IgG and IgM antibody levels were measured using the VIDAS® immunoassay system (Biomerieux, France) that is based on a two-step sandwich assay with a final florescence detection called enzyme-linked fluorescent assay. Detection of specific SAR-COV-2 IgG and IgM antibodies was performed according to the manufacturer’s instructions. Briefly, the serum samples were thawed and used immediately to minimize false results. A 100µl of serum sample were used for each test performed in duplicates. The VIDAS® SARS-COV-2 IgG and VIDAS® SARS-COV-2 IgM (Biomerieux, France) used in this study were relatively sensitive to IgG (85%–95%) and IgM (90%–95%), respectively. The index value was calculated by subtracting the relative florescence value of sera samples from the obtained florescence from the calibrator (recombinant anti-SARS-COV-2 IgG and IgM). Antibody level was expressed as one index value. A positive result is considered as ≥ 1 index value, while a negative result is considered as < 1 index value. Serum samples from 20 people who tested negative for SARS-COV-2 served as a negative control.
Statistical analysis
GraphPad Prism software (Graphpad software version 6, USA) was used for data analysis. Results are presented as the mean ± standard deviation (SD) or median. A p ≤ 0.001 was considered significant.
Ethics approval
This study was approved by the Iraqi Medical Institutions and Medical Hospitals. The consent of all patients and healthy controls was approved to conduct this study and collection of serum samples. The identity of all patients was kept confidential, as requested.
A total of 85 patients with confirmed COVID-19 symptoms were included in this study. The median age of the patients was 45 years, there were 50 men and 35 women. Symptoms were classified as moderate to severe, and some were critical cases that required mechanical ventilation for recovery. Table 1 shows the symptom profiles of the patients. A total of 67 patients (38 men and 31 women) were considered moderate cases owing to the symptoms that appeared during infection until recovery. Meanwhile, 11 patients (seven men and four women) were considered severe cases, seven of which (all male) were considered critical cases with severe symptoms and organ dysfunction and required mechanical ventilation. Unfortunately, two out of 85 patients died during this study due to organ failure and COVID-19-related complications.
Table (1):
The symptoms profile and proportions of patients in this study.
Moderate |
Severe |
Critical |
|
|---|---|---|---|
Patients number |
67 |
11 |
7 |
Age range |
44.6±5.3 |
65.3±4.1 |
69.6±3.5 |
Gender |
38 male 31 female |
7 male 4 female |
7 male 0 female |
Death |
0 |
0 |
2 male |
Patients symptoms |
High fever Shortness of breath Fatigue Cough Headache |
O2 levels lower than 80% Chest pain High level of CRP |
Failure of respiratory system Organ dysfunction Blood clots |
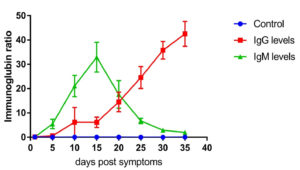
Fig. 1. IgG and IgM level profile in moderate cases. The level represents the median of 67 replicates. All patients samples were collected from day 1 until day 35 post symptoms onset with 5 days intervals. The levels of IgG and IgM were measured using VIDAS® immunoassay system (Biomerieux, France).
The IgG and IgM level profiles of patients have been reported from day 1 post-symptom onset with five-day intervals. Fig. 1 shows IgG and IgM levels in 67 moderate cases. The level of IgG was below the detection rate and considered negative on day 5, although the cases were confirmed positive using RT-PCR testing. The level of IgG increased rapidly at day 10 post symptom onset (8.232 ± 2.3, n=67) and peaked at day 35 post symptom onset (42.432 ± 4.34, n=67). IgM levels were also measured in 10 random patients with moderate cases. Moreover, IgM was detected at day 5 (6.412±2.12, n=67) and peaked at 15 days post symptom onset (32–332 ± 4.32, n=67). Afterwards, the level started to decrease sharply (1.12 ± 0.89, n=67) at day 35 post symptom onset (Fig. 1). IgG and IgM levels in patients with COVID-19 were significantly different from those in the control group (p ≤ 0.001, n=67).
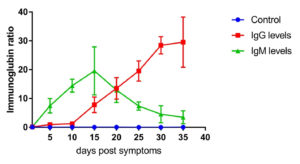
Fig. 2. IgG and IgM level profile in severe cases. The level represents the median of 11 replicates. All patients samples were collected from day 1 until day 35 post symptoms onset with 5 days intervals. The levels of IgG and IgM were measured using VIDAS® immunoassay system (Biomerieux, France).
Moreover, IgG and IgM in 11 severe cases were measured (Fig. 2). The Ig-related profile in severe cases was not relatively different from that of moderate cases. As shown in Fig. 2, IgG was detected at day 10 after symptom onset (1.323 ± 0.343, n=11). Afterwards, it started increasing gradually, reaching 29.332 ± 5.44 (n=11) on day 30 post symptoms onset. On the other hand, IgM was detected on day 5 after symptom onset (7.223 ± 3.2, n=11), peaked on day 15 post symptom onset (19.547±5.54, n=11), and then decreased sharply (3.321±2.1, n=11) on day 35 post symptom onset. IgG and IgM levels in all patients were significantly different at all time points compared to those in the control group (p≤0.001).
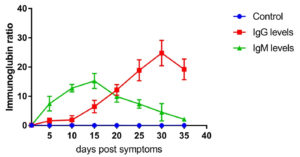
Fig. 3. IgG and IgM level profile in severe cases. The level represents the median of 7 replicates. All patients samples were collected from day 1 until day 35 post symptoms onset with 5 days intervals. Two patients were reported dead on day 20. The levels of IgG and IgM were measured using VIDAS® immunoassay system (Biomerieux, France).
Fig. 3 shows the IgG and IgM levels in critical cases from day 1 after symptom onset. IgG level was detectable from day 5 after symptom onset (1.512 ± 0.334, n=7) and increased in a time-dependent manner, peaking on day 35 post symptom onset (24.546 ± 3.22, n=5). Furthermore, the IgM response in critical cases was relatively low compared to moderate and severe cases. IgM was detectable on day 5 post symptoms (7.434 ±0.441, n=7), peaked on day 15 post symptoms onset (15.325 ± 2.42, n=5), and then sharply decreased (2.124 ± 0.434, n=5). IgG and IgM levels were significantly different between the critical and control groups (p≤0.001). For patients who have died on days 22 and 24 post symptom onset, we measured the presence of IgG and IgM until day 20.
The COVID-19 outbreak has become the most increasing pandemic since its first discovery in China in late 2019.1 Therefore, there is an urgent need for a fast diagnosis of SARS-COV-2 in patients with basic symptoms of COVID-19. Time is critical in detecting the virus as patients may not be able to combat viral invasion if it is in the late stage.12 Since the pandemic started, several approaches for the successful diagnosis of COVID-19 have been developed. The IgG and IgM level profiling has become the routine for the host humoral immune response against SARS-COV-2. Detection of IgG and IgM levels in symptomatic patients with COVID-19 has become consistent with other vital diagnostic methods, such as standard RT-qPCR and CT scans of the chest. Moreover, several detection methods to analyze immunoglobulin levels are also available, such as the lateral flow immunoassay, two step indirect immunoassay with direct chemiluminescence technology, and enzyme-linked immunosorbent assay (ELISA).22-25
It has been reported that during SARS-COV-2 infections, IgG and IgM production are consistent in patients for all disease stages. IgM is detectable on days 3–5 post viral infection, which plays a key role in the early detection of the virus.26 However, IgM may not be useful in exploring the humoral immune response during long-term defense because it decreases sharply after the patient recovers. On the other hand, IgG production was detected on days 7–10 days post viral infection. Therefore, it is important to observe the IgG profile in patients as it persists in the body after viral infection. Furthermore, some symptomatic patients with COVID-19 appear to have negative results in RT-PCR, as the technique is viral load dependent.10,17
In this study, we examined the IgG and IgM levels in patients who were confirmed to have COVID-19 and displayed symptoms. Our study showed increasing levels of IgG and IgM in a time-dependent manner. However, IgM showed a sharp decrease on day 15 after the onset of symptoms. This reflects a typical adaptive immune response against viral infection.26 Several studies also observed IgG and IgM levels in patients with COVID-19.27-31 A report by Li et al.24 showed increased IgG and IgM levels in 20 patients with confirmed COVID-19 at a relatively early stage of infection. Our results also reported that the production of IgG and IgM is dependent upon the severity of cases; in critical cases, the production of IgG and IgM was detectable as early as 5 days post symptom onset, similar to the observations of Park et al.32
Although the serological process for COVID-19 diagnosis supports other standard techniques, it has several limitations. First, the specification of anti-spike IgM and IgG might cross-react with other diseases, such as seasonal coronavirus infections and other severe infections. The presence of autoantibodies in patients with autoimmune diseases cannot be ignored, as they can be highly interfered with COVID-19 antibodies.33 However, cross-reaction with other viral infections is estimated to be very low (0.8%).34 Thus, cross-reactivity may not be relevant in our study. Nevertheless, further studies are needed to rule out the false positive results. Second, the number of case participants and serum samples might not be sufficient as some participants decided not to participate, and some have passed away during the study. The number of participants in each study group might have affected the results. Detection of anti-spike IgG and IgM may also be limited; therefore, further detection of antibodies against other viral parts is needed, such as anti-nucleoside proteins.
ACKNOWLEDGMENTS
This study is supported by the Department of Biology, College of Education for Pure Sciences, University of Mosul. Many thanks to all participants, patients to donate their blood samples. A great thanks to Central Laboratories/Nenawa Health Office including doctors and nurses who were supportive to accomplish this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the institutional ethics committee and Iraqi medical hospitals. The collection of serum samples and consent of all patients and healthy controls was approved by the health committee, and the identity of all patients was kept confidential.
AVAILABILITY OF DATA
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
- Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-733.
Crossref - Thompson RN. Novel Coronavirus Outbreak in Wuhan, China, 2020: Intense Surveillance Is Vital for Preventing Sustained Transmission in New Locations. J Clin Med. 2020;9(2):498.
Crossref - Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506.
Crossref - Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-1062.
Crossref - Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475-481.
Crossref - Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221-236.
Crossref - Ai T, Yang Z, Hou H, et al. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296(2):E32-E40.
Crossref - Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology. 2020;296(2):E41-E45.
Crossref - Bitsadze VO, Grigoreva K, Khizroeva JK, et al. Novel coronavirus infection and Kawasaki disease. J Matern Fetal Neonatal Med. 2020:1-5.
Crossref - Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133(9):1015-1024.
Crossref - Wolff F, Dahma H, Duterme C, et al. Monitoring antibody response following SARS-CoV-2 infection: diagnostic efficiency of 4 automated immunoassays. Diagn Microbiol Infect Dis. 2020;98(3):115140.
Crossref - Hou H, Wang T, Zhang B, et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunol. 2020;9(5):e01136.
Crossref - Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518-1524.
Crossref - Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323(18):1843-1844.
Crossref - Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
Crossref - Lv H, Wu NC, Tsang OT, et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020;31(9):107725.
Crossref - Qu J, Wu C, Li X, et al. Profile of Immunoglobulin G and IgM Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020, 71(16):2255-2258.
Crossref - Jiang L, Tang K, Levin M, et al. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect Dis. 2020;20(11):E276-E288.
Crossref - Catanzaro M, Fagiani F, Racchi M, Corsini E, Govoni S, Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct Target Ther. 2020;5(1):84.
Crossref - Ragab D, Eldin HS, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol. 2020;11:1446.
Crossref - Tufan A, Avanoglu Guler A, Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci. 2020;50(SI-1):620-632.
Crossref - Anderson KS, Cramer DW, Sibani S, et al. Autoantibody signature for the serologic detection of ovarian cancer. J Proteome Res. 2015;14(1):578-586.
Crossref - Che XY, Hao W, Qiu LW, et al. Antibody response of patients with severe acute respiratory syndrome (SARS) to nucleocapsid antigen of SARS-associated coronavirus. Di Yi Jun Yi Da Xue Xue Bao. 2003;23(7):637-639.
- Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349(5):508-509.
Crossref - Bai Z, Li Q, Chen Q, et al. Clinical significance of serum IgM and IgG levels in COVID-19 patients in Hubei Province, China. Journal of Intensive Medicine. 2021.
Crossref - Racine R, Winslow GM. IgM in microbial infections: taken for granted? Immunol Lett. 2009;125(2):79-85.
Crossref - Hanson KE, Caliendo AM, Arias CA, et al. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19:Serologic Testing. Clin Infect Dis. 2020.
Crossref - Shah J, Liu S, Potula HH, et al. IgG and IgM antibody formation to spike and nucleocapsid proteins in COVID-19 characterized by multiplex immunoblot assays. BMC Infect Dis. 2021;21(1):325.
Crossref - Nagasawa M, Yamaguchi Y, Furuya M, et al. Investigation of Anti-SARS-CoV-2 IgG and IgM Antibodies in the Patients with COVID-19 by Three Different ELISA Test Kits. SN Compr Clin Med. 2020;2:1323-1327.
Crossref - Sun B, Feng Y, Mo X, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):940-948.
Crossref - Feng Y. Clinical Value of SARS-CoV2 IgM and IgG Antibodies in Diagnosis of COVID-19 in Suspected Cases. J Inflamm Res. 2020;13:1089-1094.
Crossref - Park WB, Perera RA, Choe PG, et al. Kinetics of Serologic Responses to MERS Coronavirus Infection in Humans, South Korea. Emerg Infect Dis. 2015;21(12):2186-2189.
Crossref - Wang Y, Sun S, Shen H, et al. Cross-reaction of SARS-CoV antigen with autoantibodies in autoimmune diseases. Cell Mol Immunol. 2004;1(4):304-307.
- Qu JX, Gu L, Pu ZH, et al. Viral etiology of community-acquired pneumonia among adolescents and adults with mild or moderate severity and its relation to age and severity; Beijing Network for Adult Community-Acquired Pneumonia (BNACAP) BMC Infect Dis. 2015;15:89.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.