ISSN: 0973-7510
E-ISSN: 2581-690X
Opportunistic behavior of commensal bacteria during severe infection, especially of the intestine is always considered as a predisposing threat for the severely ill patient admitted to hospitals. A descriptive-analytical case-control study was carried out to detect the prevalence and risk factor analysis of fecal carriage of Carbapenem-resistant Enterobacteriaceae. Patients having fecal carriage of Carbapenem-resistant Enterobacteriaceae were considered as cases and patients who were without Carbapenem-resistant Enterobacteriaceae were taken as the control in a proportion of 1:4. Carbapenem-resistant Enterobacteriaceae (CRE) was confirmed by both conventional as well as molecular methods. Methods such as Combined Disk Test, mCIMtest, and RAPIDEC CarbaNP Test were used for phenotypic identification of CRE, Whereas Real-Time (RT)-PCR was used for the detection of bla-gene encoded for CRE (blaNDM1, blaVIM, blaKPC, blaIMP, and blaOXA48). All patients belonging to medicine wards were included in the study. we screened 436 patients during the study and found 160 patients suitable for our study, out of which 32 (20%) were cases and 128 (80%) were controls. We found a total number of 25 genes out of 16 isolates, where NDM 1 was identified in maximum numbers followed by KPC &VIM. Standard statistical analyses such as chi-squire and odd ratios were conducted to determine the risk factor of different variables. Stepwise multiple logistic regressions were carried out, where we found, that transfer from other wards to medicine ward, use of nebulizer and intravenous catheter, and use of multiple antibiotics were still statistically significant. Implementation of Multi-modal colonization prevention and control is the need for the present situation throughout the world.
Gut colonization, CRE, Fecal Carriage, NDM-1, RT-PCR
The human gut microbiome predominantly is colonized heavily by the family members of Enterobacteriaceae as Commensal and leaves no room for pathogenic bacteria to grow within. Commensal bacteria usually cause infection in healthy individuals accidentally by colonizing the sterile compartment of the body. Additionally, whenever a patient’s immune system becomes weak, the risk of opportunistic infection by these commensals increases many folds. This seriality of endogenous infection and spread by antibiotic sensitive commensal microflora is now a matter of the past. At present we have a bigger challenge to deal with; since gut colonization by resistant bacterial strains had been reported, which usually occurs by replacing gut commensal flora.1 Gut also provides the ecosystem even for antibiotic-resistant bacteria to colonize which serves as a reserve, and which subsequently may be the cause for an origin of endogenous infection and spread of such resistant bacterial strain. Antibiotic pressure, overuse of other drugs prolonged hospital stay, and immune response of patients are some of the risk factors among many factors which synergistically responsible for such colonization.2 Carbapenem-resistant Enterobacteriaceae (CRE) is one such family of bacteria, which particularly colonize the intestine.3 Gut colonization by Carbapenem-resistant Enterobacteriaceae, their endogenous spread, and exogenous cross-transmission in closed hospital settings have been reported not only in India but throughout the world.1,4
Carbapenem-resistant Enterobacteriaceae causes life-threatening infection among patients with a lower immune response which may be because of factors such as, patients with comorbidities, a patient hospitalized in Intensive Care Unit (ICU), and patients with cancer.5-7 The principal mechanism of Carbapenem-resistant is based on the production of Carbapenemase, the enzyme which inactivates Carbapenems directly8 Carbapenemase-producing CRE is distributed throughout the world and the reason for such spread is because of horizontal blagene transfer since they are on the plasmid, a mobile genetic element. Carbapenemase of global importance include Klebsiella pneumoniae Carbapenemase (KPC), New Delhi Metallo-β-lactamase type 1 (NDM-1), Verona integron encoded Metallo-β-lactamase (VIM), Imipenemase Metallo-β-lactamase (IMP), and Oxacillinase 48 (OXA-48).
Nonenzymatic Carbapenem resistance is arbitrated by a collective mechanism, generally by way of production of extended-spectrum cephalosporinase such as AmpC with decreased permeability of bacterial cell wall to the influx of Carbapenem antibiotics due to mutation of porins or maybe through the production of the extended-spectrum of β-lactamase plus decreased permeability of bacterial cell wall by Carbapenems again due to porins mutations.8 This study was conducted to broadly estimate fecal carriage of Carbapenem-resistant Enterobacteriaceae and specifically rate Carbapenemase-producing Enterobacteriaceae, and to identify possible risk factors for such acquisitions.
Study design
A prospective observational case-control study was conducted from 19/03/20 to 17/03/21.
Inclusion criteria
Patient must be aged between 13-70 years of age. Patient who remained admitted in hospital for more than 48hrs. All randomly selected inpatients of a tertiary care hospital with confirmed Carbapenem-resistant Enterobacteriaceae in stool culture admitted in medicine wards or were transferred from other wards in the medicine ward were included in the study as cases. For every one case, four randomly selected patients of the same unit found with Carbapenem-sensitive Enterobacteriaceae in their stool were taken as controls.
Exclusion criteria
Age group of patients between <13 years >70. Stool culture from patients showing growth of other bacteria than Enterobacteriaceae. Patients whosoever didn’t answer the questionnaire and whose case sheet records were unavailable, irrespective of the sample availability were excluded from the study.
Ethical status
The study was approved by the Institutional Human Ethics Committee (IHEC) and informed consent in written was obtained from individuals before they participated in the study followed by sample collection.
Sample collection and processing
Stool samples were collected after a minimum of 48 hours of hospitalization. Stool samples received routinely in the department of microbiology were also included in the study including samples from ICUs. Stool samples were received in a 5ml sterile screw-capped container. A pinch of stool samples was picked with the help of an inoculating loop and emulsified in 2ml of sterile 0.85% normal saline.9 Samples were sequentially cultured on MacConkey agar plate and the colonies were further processed through a battery of primary microbiological tests such as gram staining, motility testing, and catalase, as well as biochemical test to identify the isolate up to species level.
Antibiotic susceptibility testing
Preliminary identification of CRE was carried out employing antibiotic susceptibility testing (AST) carried out as per CLSI (2017) guidelines by using antibiotic discs of Gentamicin, Amikacin, Ciprofloxacin, Cotrimoxazole, Cefotaxime, Piperacillin-Tazobactum, Imipenem, Meropenem, Tigecycline, Colistin, Polymyxin B, and Fosfomycin (Himedia). Control strains used were E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923.
Phenotypic procedures
Carbapenem-resistant Enterobacteriaceae were also determined here by specific methods recommended by Clinical and Laboratory Standard Institute (CLSI 2017) such as mCIMTest and RAPIDEC Carba NP Test, and Double Disk Synergy Test.
Combined Disc Test
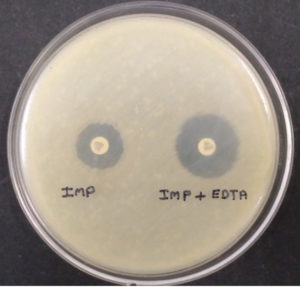
Ethylene-diamine-tetra-acetic acid (EDTA) is an amino-poly-carboxylic acid that binds metal ions. The same metal chelating properties of EDTA was used here for phenotypic detection of Metallo-Beta-Lactamases (MBLs) producing bacterial isolates as it can also chelate zinc and degrade MBLs. The test was carried out as described by Khosravi Y et al. The test isolates grown on blood agar were inoculated in peptone water for four hours, when the culture was in log phase, it was spread over on Mueller Hinton agar plate. Then two Imipenem discs were placed on the plate at a distance of 26mm from each other, followed by the addition of 10µl of 750µl/ml EDTA solution on one. The zone of inhibition was compared after overnight incubation at 37°C.10 The difference of ≥7mm between the two (Imipenem and Imipenem with EDTA) was considered to be positive for the Test. EDTA is not water-soluble, so to dissolve EDTA in water NaOH was used to maintain the pH 8.
RAPIDEC® CARBA NP Test
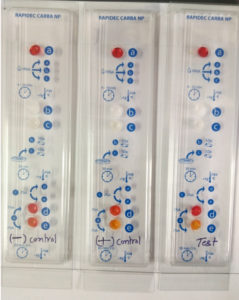
The test detects Carbapenemase enzymes in Enterobacteriaceae. The test is based on the detection of the hydrolysis of the β-lactam ring of Imipenem.11 The test procedure consists of three steps of a CARBA NP test kit, 1st step – dehydration where 100µl of suspension medium were added to wells a, b, and c, 2nd step – lysis where test colonies were inoculated in well c and compared its turbidity with well b, and 3rd final step- hydrolysis where phenol red indicator from well a and test suspension from well c was added together to both, control well d and test well e which contains Imipenem. Making control well d as (phenol red + bacterial Lysate) and test well e as (phenol red + bacterial Lysate + Imipenem). Hydrolysis of Imipenem by bacterial Lysate which is detected as by change in pH value by using indicator phenol red. And the interpretation was made by comparing the control well d with test well e. if a color change occurs from red to yellow, or light orange the result is positive (Fig. 2). The test was developed by BioMerieux (SA, France) and was approved and recommended by the Centers for Disease Control and Prevention (CDC), The European Committee on Antimicrobial Susceptibility Testing (EUCAST), and The Clinical & Laboratory Standards Institute (CLSI).12
mCIM Test
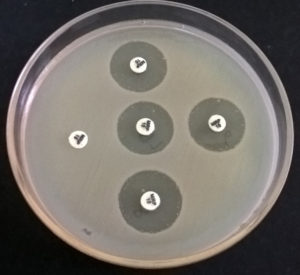
The test detects Carbapenemase Producing-Carbapenem Resistant Enterobacteriaceae (CO-CRE) from Non- Carbapenemase Producing-Carbapenem Resistant Enterobacteriaceae, which may be due to cumulative effect of other factors such as Extended Spectrum of β-Lactamases or Extended-spectrum cephalosporinase and decreased permeability (e.g., porin mutations) of the bacterial cell. The test is recommended by CLSI as well as CDC. In the modified-CIM test, a 10µg Meropenem disc was immersed and incubated with test strain in nutrient broth (0.5 McFarland opacity) for up to 4 hrs. Then E. coli ATCC 25922 strain was also used to prepare bacterial suspension similar to the turbidity of 0.5 McFarland standards and spread over to Mueller Hinton Agar plate. Subsequently, the meropenem disc was placed on the MH Agar plate and incubated overnight. Test strain with a zone of inhibition <16mm was considered to be positive, whereas the zone of inhibition ≥ 19 was considered negative.13
Real-time qPCR to detect the gene for carbapenemase-producing CRE
DNA was isolated by using HiPurA® Genomic DNA Purification kits (Hi-Media®, India) which is based principally on reversible nucleic acid binding properties of the advanced silica gel membrane and the speed plus the versatility of Miniprep spin columns to yield high quality of DNA. The method contains three basic steps. 1st adsorption of test DNA to the membrane where log-phase bacterial culture was harvested by centrifugation and lysed by lysozymes, and then allowed to bind to the silica-gel membrane of the HiEluteMiniprep spin column. 2nd step removal of residual contaminants as a contaminant was made a pass through the membrane and finally 3rd step, where pure genomic DNA was eluted and collected in a tube. Primers used for the identification of carbapenem-resistant genes were of global importance.14-15 All five primers were previously designed and synthesized by Helini biomolecules, Chennai. The primers were as follows blaNDM (forward primer-GCAGCACACTTCCTATCTCG and reverse primer-GTCCATACCGCCCATCTTGT a total 202bp size), blaKPC (forward primer-CGGCAGCAGTTTGTTGATTG and reverse primer- CGCTGTGCTTGTCATCCTTG a total of 275bp size.), blaVIM (forward primer- TCCGTGATGGTGATGAGTTG and reverse primer-CGCTCGATGAGAGTCCTTCT a total of 115bp size), blaIMP(forward primer-GCTTGATGAAGGCGTTTATGTT and reverse primer-GCCGTAAATGGAGTGTCAATTAG a total of 387bp size), and blaOXA forward primer-CTGAACATAAATCACAGGGCGT and reverse primer- GAAACTGTCTACATTGCCCGAA a total of 360bp size).
Real-time UniplexqPCR was performed using Sso Advanced in Universal SYBR® Green Supermix by BioRad, which contains antibody-mediated hot start Sso 7d fusion polymerase, dNTPs, MgCl2, SYBR® General Dye, enhancer, stabilizers, and a blend of passive reference dye including ROX and fluorescein. All components of master mix, DNA template, forward primers, reverse primers, and molecular grade water was used in a volume of 10 µl, 3 µl, 2 µl, 2 µl, and 3 µl, respectively. The total reaction volume for qPCR was 20 µl and a BioRad thermal cycler was used to carry out the tests. The protocol followed is as follows – polymerase activation and DNA denaturation-3 minutes at 98°C, denaturation for 15 seconds at 98°C, and annealing/extension and plate reading at 60°C for 30 seconds. Step 2nd (denaturation and extension) was repeated for 35 cycles. A melt curve was observed at 65-95°C, 0.5°C increment at 2-5sec/step. Using instrument default setting. All PCR reaction was performed using positive control for each gene and RNase free water as a negative control (Non-Template Control).16
Statistical analysis
Numerical variables were performed by measuring variability and central tendency, followed by calculation of median and Interquartile range. Categorical variables were measured by their frequencies and their proportion for every category. Univariate analysis was conducted to calculate the significant risk factor. P-value of 0.5 was used as a level of significance for testing the null hypothesis. Stepwise multiple logistic regression analyses of the risk factor for colonization by CRE were also calculated for variables which were found significant on standard statistical analysis to identify independent risk factors. All data were analyzed by IBM SPSS STATISTICS (version 20.0, New York, USA).17
Microbiology data
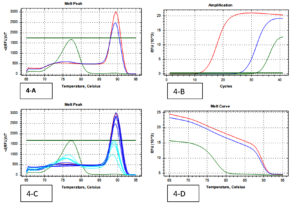
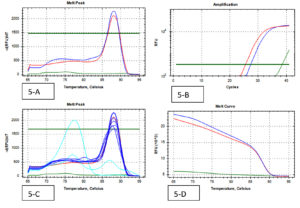
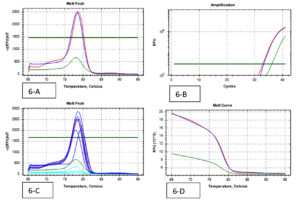
A total of 202 isolates from the family Enterobacteriaceae were acquired from 160 fecal samples, because many fecal samples had more than one islolates. Among that, the split-up of 38 CRE isolates were as follows; E. coli was the dominant species among CRE (cases) positive Enterobacteriaceae with 16 isolates, Klebsiella spp. with 17 and Enterobacter spp. were the least isolated species with 5 isolates. A similar pattern was observed in the Carbapenem Sensitive Enterobacteriaceae (controls), where 164 isolates were identified from 128 stool samples. 38 CRE isolates were identified by phenotypic methods, where 36 isolates were identified by the Combined Disc test (Fig. 1), 32 isolates were identified by RAPIDEC® CARBA NP test (Fig. 2) and only 30 were identified by mCIM test (Fig. 3). From 16 isolates the specific gene was identified by genotypic technique. Among the genotypic methods number of NDM 1 gene were 14 in numbers followed by KPC with 6 in numbers and VIM with 4 in numbers (Table 1). Real-time PCR amplification curve, melt peak, melt curve for NDM, KPC, and VIM are in Fig. 4, Fig. 5, and Fig. 6, respectively. The total numbers of a gene identified among E. coli were as follows; three isolates each with only one NDM 1, one isolate with NDM 1 and KPC, one isolate with NDM and VIM, and one isolate with three genes that is NDM 1, KPC, and VIM. Whereas the total number of a gene identified in Klebsiella spp. isolates are as follows; five isolates with NDM 1, three isolates with only KPC, one isolate with NDM 1 and VIM, and one isolate with three genes (NDM 1, KPC, and VIM). In the case of Enterobacter spp. one isolate was found with NDM 1and the other one was found with VIM.
Table (1):
Phenotypic and molecular characterization of CRE isolates.
| Number of Isolates | ||||
|---|---|---|---|---|
| Type of Tests | E.coli | K.pneumoniae | E.cloacae | Total |
| CRE by phenotypic methods | 16 | 17 | 5 | 38 |
| Combined Disc Test | 15 | 17 | 4 | 36 |
| RAPIDEC® CARBA NP Test | 13 | 15 | 4 | 32 |
| mCIM Test | 11 | 17 | 2 | 30 |
| CRE by genotypic methods | 6 | 8 | 2 | 16 |
| blaNDM1 | 6 | 7 | 1 | 14 |
| blaKPC | 2 | 4 | 0 | 6 |
| blaIMP | 0 | 0 | 0 | 0 |
| blaVIM | 2 | 2 | 1 | 5 |
| blaOXA-48 | 0 | 0 | 0 | 0 |
Fig. 1. (Combined Disc Diffusion Test -Combined Imipenem + EDTA shows enhanced zone of inhibition ≥7mm as compared to Imipenem alone indicates positive tests.
Fig. 2. (RAPIDEC®CARBA NP Test– development of orange color in well (e) as compared to well (d) indicates a positive test).
Fig. 3. (mCIM Test- in the above picture only one sample with completely no zone or inhibition by meropenem is positive)
Analysis of CRE case vs. CSE controls
When CRE cases were compared with CSE controls for current hospitalization related factors, univariate analysis in Table 2 showed that cases were more likely male. The length of hospital stay before sample collection in cases was higher with an Interquartile range (IQR) of 120 hours (5 days) while it was 72 hours (3 days) for controls. The potential risk factors for CRE colonization are as follows; use of Intra Venous Catheter (P-value 0.000), patient with Anemia (P-value 0.000), Patient with Diarrhea (P-value 0.014) and Diabetes mellitus (P-value 0.022), use of cefotaxime (P-value 0.000), ciprofloxacin (P-value 0.000), Albendazole (P-value 0.001) and Rantac (P-value 0.004) and use of multiple antibiotics for a single patient (P-value 0.034).
Table (2):
Univariate analysis of current hospitalization-related factors associated with faecal carriage of Carbapenem resistant Enterobacteriaceae (Cases- CRE isolates from patient gut/ Control- CSE isolates from patient gut).
Characteristics (variables) |
Cases (N=32) |
Controls (N=128) |
Odds Ratio |
(95%CI) |
P-value (chi-square) |
|---|---|---|---|---|---|
Age |
0.97 |
0.95-1.00 |
0.088 |
||
Median |
47.50 |
37.50 |
|||
Interquartile range |
41.25-60.0 |
30.50-57.0 |
|||
Sex (Male) |
26 |
95 |
1.505 |
0.569-3.979 |
0.407 |
LHS* before CRE isolation |
0.91 |
0.85-0.94 |
0.063 |
||
Median |
120 |
72.06 |
|||
Interquartile range |
120-144 |
60-90 |
|||
Intensive care unit |
3 |
5 |
2.545 |
0.575-11.263 |
0.204 |
TFW** |
6 |
3 |
9.615 |
2.258-40.945 |
0.000 |
Smoking |
2 |
8 |
1.000 |
0.202-4.955 |
1.000 |
Alcoholic |
2 |
7 |
1.152 |
0.228-5.832 |
0.864 |
Nebulizer |
8 |
27 |
1.247 |
0.504-3.085 |
0.633 |
Intravenous catheter |
24 |
24 |
13.00 |
5.207-32.458 |
0.000 |
Anemia |
14 |
13 |
6.880 |
2.787-16.986 |
0.000 |
Fever |
6 |
26 |
0.905 |
0.338-2.428 |
0.843 |
Diarrhea |
6 |
7 |
3.989 |
1.238-12.850 |
0.014 |
Diabetic Mellitus |
10 |
18 |
2.778 |
1.131-6.821 |
0.022 |
Azithromycin |
2 |
9 |
0.881 |
0.181-4.295 |
0.876 |
Cefotaxime |
16 |
20 |
5.400 |
2.328-12.525 |
0.000 |
Ceftriaxone |
2 |
20 |
0.360 |
0.080-1.628 |
0.168 |
Ciprofloxacin |
12 |
9 |
7.933 |
2.961-21.256 |
0.000 |
Albendazole |
10 |
12 |
4.394 |
1.691-11.418 |
0.001 |
Metronidazole |
4 |
8 |
2.143 |
0.603-7.621 |
0.230 |
Emeset |
6 |
23 |
1.054 |
0.389-2.852 |
0.918 |
Ranitidine |
16 |
31 |
3.129 |
1.403-6.980 |
0.004 |
Pantoprazole |
8 |
18 |
2.037 |
0.794-5.229 |
0.134 |
Multiple antibiotics |
4 |
3 |
5.952 |
1.261-28.101 |
0.034 |
LHS*Length of Hospital Stay
TFW**Transferred From other Wards
CRE- Carbapenem Resistance Enterobacteriaceae
CSE- Carbapenem Sensitive Enterobacteriaceae
Note: Variables that were not able to be statistically analyzed (remains constant) are not in the list (vegetarian, Amikacin, Amoxicillin clavulanic acid, Inguinal Hernia, Hemorrhoids, PiperacillinTazobactam, cefoperazone, and levofloxacin).
Fig. 4. (Real-Time PCR curves of NDM 1- Red lines-positive control, Green lines-negative control, Blue lines-positive samples & Light blue negative samples).
Fig. 5. (Real-Time PCR curves of KPC – Red lines-positive control, Green lines-negative control, Blue lines-positive samples & Light blue negative samples).
Fig. 6. (Real-Time PCR curves of VIM – Red lines-positive control, Green lines-negative control, Blue lines-positive samples & Light blue negative samples).
A significant odd ratio has been found for the variable are as follows; patient transferred from other wards (odd ratio: 9.6), patient with the intravenous catheter (odd ratio: 13.0), and patient taking ciprofloxacin (odd ratio: 7.9) on standard statistical analysis. Stepwise multiple logistic regression of significance variable on standard method, reasserted the findings where a limited number of variables are still found to be significant as mentioned in Table 3, as independent risk factors.
Table (3):
Stepwise multiple logistic regressions of significant variables on the statistical standard method.
| B | S.E. | Wald | Sig. P-value | Exp(B) odd ratio | 95% C.I.for EXP(B) | |||
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Step 5e | Intravenous catheter | 2.020 | .580 | 12.133 | .000 | 7.541 | 2.419 | 23.503 |
| TFW** | 1.799 | .840 | 4.590 | .032 | 6.043 | 1.166 | 31.328 | |
| Cefotaxime | 1.268 | .532 | 5.684 | .017 | 3.553 | 1.253 | 10.077 | |
| Ciprofloxacin | 1.330 | .662 | 4.031 | .045 | 3.781 | 1.032 | 13.853 | |
| Albendazole | 2.072 | .645 | 10.318 | .001 | 7.943 | 2.243 | 28.126 | |
In the present scenario, CRE has emerged as a major public health care pathogen and is responsible to cause infection during hospitalization even in our study. The fecal carriage of CRE makes it one of the persistent potential risk factor which can turn down the overall recovery of the patient in hospitals. Therefore routine surveillance of fecal carriage must be recommended and adopted by infection control policymakers in India.
In the western world, fecal carriage surveillance is a common practice and is recommended by almost all agencies such as the Center for Disease Control and Prevention (CDC), National Services Scotland (NHS), Australian Commission on Safety and Quality in Health Care, European Center for Disease Control and Prevention (ECDC) and many more. In India, very few studies so far have been conducted to assess the prevalence and incidence of fecal colonization. A study conducted by Mohan et al. Where he reported the prevalence of CRE fecal carriage at 18.1%, while another study conducted by Rai et al. reports CRE prevalence at 9.9%, from a similar study.19
As per our knowledge, this is the 1st case-control study to identify and assess the risk factors for fecal carriage of CRE in India. Our findings are as follows; phenotypically mCIM test was found to be the most specific method and the combined disc diffusion test was a more sensitive method, while Rapidec carbaNP was instituted to be equally sensitive and specific among phenotypic tests. CRE positive isolates by genotypic methods were taken as a standard to say isolates sensitivity and specificity because molecular analysis always is considered very specific and sensitive. We have identified blaNDM 1 14(87.5%) the maximum, followed by blaKPC 6 (37.5%), and blaVIM 5 (31.2%). This may be because NDM1 seems to have been reported even from water sources in India.20-21
On standard statistical analysis, our result showed that there was a significant association between fecal carriage of CRE and patient using Intra Venous Catheter, patient with Anemia, Diarrhea and Diabetes mellitus, the patient using cefotaxime, ciprofloxacin, Albendazole and Rantac, and patient using multiple antibiotics at the same time.
Our variables under study i.e. patient on Intravenous catheter, Patient transferred from other wards, the patient taking cefotaxime, ciprofloxacin, and Albendazole was still found to be significant by stepwise multiple logistic regression, which makes them as independent variables for CRE gut colonization during hospitalization. Use of IVC, the patient taking ciprofloxacin and patient on multiple antibiotics, and patients on any device have been reported earlier as significant risk factors for CRE fecal colonization.19
Many research studies have established the fact that drugs especially proton pump inhibitors, antacids, oral hypoglycemic such as metformin, antibiotics, and laxatives have associated with the decolonizing of the gut.22-23. Decolonization of the gut by any means is fundamentally required for colonization of the same by other favored or unfavored pathogens.
The present case-control study also calculates the odds ratio of risk factors associated with fecal carriage i.e. use of intravenous catheter topped the chart with an odds ratio of 13.0 on standard statistical analysis and the odds ratio of 7.5 on logistic regression.
Which concludes; the risk factor of CRE carriage in a patient with IVC was 13 times higher than a non-IVC user-patient by standard statistical analysis, whereas the risk factor of CRE carriage in a patient with IVC was 7.5 times higher than a non-IVC user-patient even as an independent risk factor.
Similarly, the risk of CRE colonization was found to be 9.0 times and 6.0 times higher in the patient who was transferred from other wards to medicine wards, by the standard statistical method and by logistic regression method, respectively.
Multiple Carbapenem hydrolyzing gene such as blaIMP-1, and blaVIM-1 genes besides blaOXA-51 in Acinetobacter baumannii have already been reported.24 Even multiple Carbapenemase resistance genes in isolates of Enterobacteriaceae have been reported from a study conducted in Iraq.25 Even we also found multiple Carbapenemase genes in our isolates which are used to be a common finding in current scenario. Multi-Drug Resistance (MDR) bacterial strains use to have multiple resistance genes as well as uses multiple resistance mechanisms.
So, implementation of multi-modal colonization prevention policies i.e. hand hygiene compliance, surveillance cultures for asymptomatic CRE carriers, contact precaution, patient isolation, and environmental cleaning should be recommended, which can play a significant role in controlling fecal carriage of CRE
Our study is the first case-control study reporting in the same settings on risk factors of CRE, including CPE and non-CPE in India. Beyond identifying the gene encoding for the commonly disseminated carbapenemase-producing Enterobacteriaceae among inpatient, it evidences, the significance of the common risk factors associated with fecal carriage of CRE on hospitalization. The study result highlights the importance of antibiotic stewardship, selective approach on the prescription of other drugs, reducing duration of hospital stay, taking effective patient isolation measures, and correct cleaning and disinfection procedures as per universal guidelines to avoid the spread of CRE in hospital settings. Colonization control measures are as important as infection control measures, as no one knows how readily one’s colonization can become one’s infection. Knowledge of risk factors and implementation of control measures such as reducing the length of patient stay at the hospital, effective time-bound surveillance of gut colonization could play a devise role in the spread of CRE, even spread of other resistant bacterial strains in hospitals.
ACKNOWLEDGMENTS
The authos would like to thank Dr. A. John William Felix, Reader Cum Statistician, Department of Community Medicine, Rajah Muthiah Medical College, Annamalai University, for his role in statistical computations. We also thank teaching and nursing staffs of the Medicine Department, World College of Medical Science; Research and Hospital, for their cooperation and constant support throughout the study.
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
R reviewed the literature, conducted tests, and drafted of manuscript. PK designed Tables and charts. MK, PR and MJ supervised and reviewed the manuscript. All authors read and approved the manuscript.
FUNDING
None.
ETHICS STATEMENT
The study was approved by the Institutional Human Ethics Committee (IHEC), Rajah Muthiah Medical College, India with reference number IHEC/0371/2018.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Gorbach SL. Microbiology of the Gastrointestinal Tract. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 95.
- Bharadwaj R, Robinson ML, Balasubramanian U, et al. Drug-resistant Enterobacteriaceae colonization is associated with healthcare utilization and antimicrobial use among inpatients in Pune, India. Bharadwaj et al. BMC Infect Dis. 2018;18(1):504.
Crossref - K A Young, L J Se, P S Yoon, et al., Risk Factors for Carbapenemase-Producing Enterobacterales Infection or Colonization in a Korean Intensive Care Unit: A Case–Control Study. Antibiotics 2020, 9, 680.
Crossref - Nicolas-Chanoine MH, Vigan M, Laouenan C, Robert J, on behalf of the BE-carb Study Group. Risk factors for carbapenem-resistant Enterobacteriaceae infections: a French case-control-control study. Eur J Clin Microbiol Infect Dis. 2019;38(2):383-393.
Crossref - Ravikant, Jeya M, Felix AJW. Prevalence and risk factor analysis of fecal carriage of carbapenem-resistant Enterobacteriaceae a Tertiary Care Hospital. J Clin Diagn Res. 2019;13(7):DC01-DC06.
Crossref - Mittal G, Gaind R, Kumar D, et al. Risk factors for fecal carriage of carbapenemase-producing Enterobacteriaceae among intensive care unit patients from a tertiary care center in India. BMC Microbiol. 2016;16(1):138.
Crossref - Jaiswal SR, Gupta S, Kumar RS, et al. Gut Colonization with Carbapenem-resistant Enterobacteriaceae Adversely Impacts the Outcome in Patients with Hematological Malignancies: Results of A Prospective Surveillance Study. Mediterr J Hematol Infect Dis. 2018;10(1):e2018025.
Crossref - Washington State Department of Health. Carbapenem-resistant Enterobacteriaceae Reporting and surveillance guidelines. DOH. 2018:420-097:1-13.
- Abdallah MH, Alnaiemi N, Reuland EA, et al. Fecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital-based cross-sectional study. Antimicrob Resist Infect Control. 2017;6:62.
Crossref - Khosravi Y, Loke MF, Chua EG, Tay ST, Vadivelu J. Phenotypic Detection of Metallo-β-Lactamase in Imipenem-Resistant Pseudomonas aeruginosa. The Scientific World Journal. 2012;2012:654939.
Crossref - Pierce VM, Simner PJ, Lonsway DR, et al. Modifiedcarbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55:2321-2333.
Crossref - Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503-1507.
Crossref - Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 27thInformational Supplement; (2017) M100-S27. Wayne, PA, USA.
- Monteiro J, Widen RH, Pignatari ACC, Kubasek C, Silbert S. Rapid detection of carbapenemase gene by multiplex real-time PCR. J Antimicrob Chemother. 2012;67(4):906-909.
Crossref - Choudhury DD, Singh NP, Rai S, Batra P, Manchanda V. Carbapenem-resistant Enterobacteriaceae neonatal gut colonization: A future concern in healthcare settings. Indian J Microbiol Res. 2018;5(3):348-354.
Crossref - Casaburi G, Duar RM, Vance DP, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8:131.
Crossref - Lavagnoli SL, Bassetti BR, Kaiser TDL, Kutz KM, Cerutti C Jr. Factors associated with the acquisition of carbapenem-resistant Enterobacteriaceae. Rev Latino-Am Enfermagem. 2017;25:e2935.
Crossref - Karuniawati A, Saharman YR, Lestari DC. Detection of carbapenemase encoding genes in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumanii isolated from patients at Intensive Care Unit Cipto Mangunkusumo Hospital in 2011. Acta Med Indones. 2013;45:101-106.
- Mohan B, Prasad A, Kaur H, Hallur V, Gautam N, Taneja N. Fecal carriage of carbapenem-resistant Enterobacteriaceae and risk factor analysis in hospitalized patients: A single-center study from India. Indian J Med Microbiol. 2017;35(4):555-562.
Crossref - Kapil A. The challenge of antibiotic resistance needs to contemplate. Indian J Med Res. 2005;3:83-91.
- Lamba M, Graham DW, Ahammad SZ. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ Sci Technol. 2017;51(23):13906-13912.
Crossref - Vila AV, Collij V, Sanna S, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11(1):362.
Crossref - Enright EF, Gahan CGM, Joyce SA, Griffin BT. The Impact of the Gut Microbiota on Drug Metabolism and Clinical Outcome. The Yale Journal of Biology and Medicine. 2016;89:375-382.
- Niranjan DK, Singh NP, Manchanda V, Rai S, Kaur IR. Multiple carbapenem hydrolyzing genes in clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol. 2013;31:237-241.
Crossref - Gatya Al-Mayahie SM, Al-Guranie DRT, Hussein AA, Bachai ZA. Prevalence of common carbapenemase genes and multidrug resistance among uropathogenic Escherichia coli phylogroup B2 isolates from outpatients in Wasit Province/ Iraq. PLoS ONE. 2022;17(1):e0262984.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.