ISSN: 0973-7510
E-ISSN: 2581-690X
Multidrug resistance has been increasing world wide amongst most microrganisms, and adding to increased rate of both hospital and community acquired infections. Of all resistance mechanisms the alarming spread of carbapenemase producers is most worrisome and needs to be tackled head on. The present study was undertaken with the objective of determining the prevalence of carbapenemase producers and its significance in selecting the appropiate antibiotic for clinical use.The study was undertaken by the department of Microbiology and Immunology of SGRRIM&HS, Dehradun over a period of six months. A total of 1918 varied clinical specimens were subjected to Bacterial identification and antibiotic sensitivity determination. Further carbapenemase production was detected phenotypically using modified carbapenemase inactivation method (mCIM) for randomly selected 152 carbapenem resistant gram negative isolates. Total of 58.55% isolates tested mCIM test positive of which the highest percentage (71.4%) were Pseudomonas spp, while 17.2% isolates were not found to be carbapenemase producers i.e mCIM negative. These results substantiate the importance of differentiating the carbapenemase producers from non producers to aid in rational use of antibiotics.
Carbapenemase, Multidrugresistant
Multidrug resistance amongst most bacterial species has been increasing at an alarming rate adding to increased rate of both hospital and community acquired infections. One of the most significant and worrisome resistance traits is resistance to carbapenems, which more often than not constitute the last line of defense against the most lethal of gram negative bacteria This is being observed very frequently in many gram negative isolates associated with both nosocomial and some community acquired infections.
Gram negative bacteria pocessing resistance to carbapenems have been reported from all corners of the world. The resistance could be the result of either decreased outer membrane permeability combined with hyperexpression of betalactamases pocessing week carbapenemase activity or due to pocession of carbapenemase gene itself1-3. Those pocessing genes for carbapenemase production are more problematic because genetic traits are transferable both inter and intraspecies which is not the case with former where the resistance characteristic is non transferable. The Carbapenem-hydrolysing β-lactamases such as KPC, VIM, IMP, NDM and OXA-48 types are the most potent of all β-lactamases with the ability to hydrolyse almost all β-lactams. Besides these enzymes carry several resistance genes, conferring resistance to many other classes of antibiotics apart from carbapenems including other aminoglycosides, fluoroquinolones, tetracyclines, trimethoprim, sulphonamides and phenicols4,5.
Therefore it becomes imperative to differentiate carbapenemase producing strains from those where resistance is due to other mechanisms. This would not only aid in selecting the appropriate antibiotic but also containing the spread of cabapenemase resistant strains.
Various phenotypic and molecular methods are available for carbapenemases detection of which CLSI recommends three namely Modified Hodje test, carba NP and mCIM. In this study we have used mCIM for detection of carbapenemases production amongst gram negative isolates showing raised MIC for carbapenems10,12.
Aims & Objective
The study was undertaken to determine the prevalence of carbapenem resistance amongst gram negative isolates and further detection of carbapenemase production amongst these isolates using modified carbapenem inactivation method (mCIM).
The study was carried out for a period of six months from August 2019 to February 2020 by the department of Microbiology and Immunology of SGRRIM&HS, Dehradun.
Ethical clearance was obtained from the institutional ethics committee and participants privacy and confidentiality was protected for all samples.
Type of study
This was a prospective cross-sectional study. A total of 1918 varied clinical specimens collected from patients presenting to different specialties (both IPD & OPD) in the hospital were submitted to the microbiology department for aerobic bacterial isolation and antibiotic sensitivity determination.
Bacterial identification and antibiotic sensitivity was determined by automated method using VITEK 2 (Biomeriux, France). A total of 1499 gram negative isolates were detected.
Further phenotypic detection of carbapenemase production using modified carbapenemase inactivation method (mCIM) was done for randomly selected 152 gram negative isolates.
These included the three most commonly isolated gram negative species namely E.coli (n=72), Klebsiella spp (n=65) and Pseudomonas (n=14).
mCIM testing
Using a sterile inoculating loop, 1 µl of test organism was added into a tube containing 2 mL of tryptic soy broth (TSB; HiMedia) the bacterial suspension was vortexed for 10 to 15 seconds. Next, a 10-µg MEM disk (HiMedia Susceptibility Test Disc) was aseptically added into the bacterial suspension. The tube was then incubated for 4 hours ± 15 minutes at 35°C ± 2°C in ambient air. Just prior to completion of the 4-hour carbapenem inactivation step, a suspension of the mCIM indicator organism (E. coli ATCC 25922, a carbapenem-susceptible strain) with turbidity equivalent to a 0.5 McFarland standard was prepared and the surface of a MHA plate (HiMedia Mueller-Hinton Agar) was inoculated using the procedure for standard disk diffusion susceptibility testing The MEM disk was then removed from the TSB bacterial suspension using a 10-µl inoculating loop. The loop was dragged along the edge of the tube during removal to remove excess liquid, and the disk was placed onto the inoculated MHA plate, which was then incubated in an inverted position for 18-24 hours at 35°C ± 2°C in ambient air6.
mCIM result interpretation
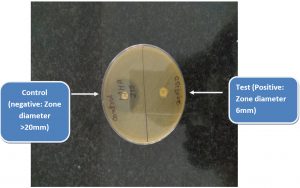
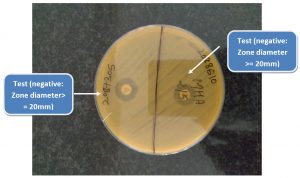
The diameter of the zone of inhibition around each MEM disk was measured (Fig. 1 & 2).
A zone diameter of 6-10 mm was considered to be a positive result (i.e., carbapenemase production detected), 11-19 mm an indeterminate result, and ≥ 20 mm a negative result (i.e., no carbapenemase production detected).A narrow ring of growth abutting the MEM disk, representing carryover of the test organism from the TSB, was ignored6. Additionally a control disk was used which had not been dipped in the bacterial suspension.
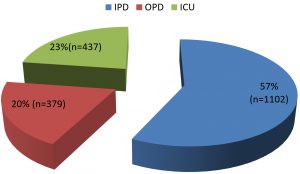
The study was conducted by the Department of Microbiology and Immmunology of Shri Mahant Indresh hospital Dehradun over a period of six months. During this period a total of 1918 samples were received from various locations in the hospital (Chart 1). Of all the samples received majority were urine (36.4%) closely followed by pus at 30.9%. The distribution of samples is depicted in Table 1.
Table (1):
Type of samples.
Sample |
Number |
Percentage |
|---|---|---|
Urine |
700 |
36.4 |
Pus |
594 |
30.9 |
Blood |
267 |
13.9 |
Tips and aspirates |
236 |
12.3 |
Fluid |
88 |
4.5 |
Others |
33 |
1.7 |
Total |
1918 |
100 |
A total of 1758 bacilli were isolated from these samples of which 1499 were gram negative bacilli (Table 2).As show in the Table 2 the most common isolate was E.coli (35%) followed by Klebsiella spp and Pseudomonas spp at 13% each.
Table (2):
Distribution of organisms.
Organisms |
Number |
Percentage |
|---|---|---|
E.coli |
624 |
35.8 |
Klebsiella spp |
235 |
13.4 |
Pseudomonas spp |
222 |
12.7 |
Acinetobacter spp |
212 |
12.1 |
Staphylococcus aureus |
101 |
5.8 |
Enterococcus spp |
87 |
4.9 |
Enterobacter spp |
81 |
4.6 |
Candida spp |
57 |
3.2 |
Proteus spp |
56 |
3.2 |
Salmonella spp |
38 |
2.1 |
Serratia spp |
28 |
1.6 |
CONS |
10 |
0.5 |
Streptococcus pyogenes |
4 |
0.2 |
Shigella spp |
3 |
0.1 |
Total |
1758 |
|
Gram Negative Isolates |
1499 |
Antimicrobial susceptibility testing was performed for the three most commonly isolated organisms using Vitek 2 (Biomerieux, France) automated system based on CLSI guidelines 2019.
Isolate showing raised MIC for either Meropenem/Imipenem or both were considered to be Carbapenem resistant as per MIC. Highest level of resistance was reported for Klebsiella spp at 58.72% followed by Pseudomonas spp at 36.5% while Escherichia.coli reported least resistance at 27.4% (Table 3).
Table (3):
Carbapenem resistance based on MIC.
Organism |
Total |
Carbapenem resistance (increased MIC) |
Percentage |
|---|---|---|---|
E.coli |
624 |
171 |
27.4 |
Klebsiella spp |
235 |
138 |
58.72 |
Pseudomonas spp |
222 |
81 |
36.5 |
Further randomly 152 of these three isolates were selected for testing of carbapenemase production using modified carbapenem inactivation test (mCIM). Seventy two isolates of E.coli, 65 isolates of Klebsiella spp and 14 isolates of Pseudomonas were tested(Table 4).
Table (4):
Modified Carbapenemase Inactivation method (mCIM).
Organism |
Tested |
Positive |
Percentage |
|---|---|---|---|
E.coli |
72 |
42 |
58.33 |
Klebsiella spp |
65 |
37 |
56.92 |
Pseudomonas spp |
14 |
10 |
71.42 |
Total |
151 |
89 |
58.94 |
As shown in the Table 4 a total of 58.94% ( n=89/151) isolates tested positive for (mCIM test).Of these highest percentage (71.4%; 10/14) were Pseudomonas spp followed by E.coli (58.33%;n=42/72) and Klebsiella spp (56.9 %;37/65).
Table 5 elaborates the carbapenem profile of the one hundred and fifty two isolates that were subjected to modified carbapenemase inactivation method.
Table (5):
Carbapenem profile of isolates.
Resitance profile |
Number |
Percentage |
|---|---|---|
MIC raised only |
26 |
17.2 |
mCIM only |
23 |
15.3 |
MIC+mCIM |
68 |
44.8 |
Sensitive |
35 |
23 |
Total |
152 |
100 |
As shown in the table, 45% showed both raised MIC as well as evidence of carbapenemase production while,17% showed raised MIC (for carbapenems) alone This implies that in these isolates carbapenem resistance is due to mechanism other than carbapenemase production and they are likely to be sensitive to one or more carbapenem (barring the carbapenem for which MIC is raised).
In our study a total of 1499 gram negative isolates were obtained of which the predominant gram negative bacilli was E.coli (35%). This has also been reported in another similar study by Dahab et al. where also E.coli was the most common GNB isolated (54.4%)7.
Authors from various parts of India have reported varying resistance rates of carbapenem in Enterobacteriaceae ranging from 5.75 % to 51% in various gram negative isolates (8,9).In our study raised MIC for carbapenems was detected in 27.4% (171/624) of E.coli, 58.72%(138/235) of Klebsiella and 36.5% (81/222) of Pseudomonas. These findings have also been reported by Sathya Pandurangan et al. where in they have reported resistance in E.coli at 31%, and Klebsiella at 51%9.
In this study 36 % of the isolates showed decreased susceptibility to carbapenems via raised MIC using automated method. This was is agreement with another study by Hayajneh WA11 wherein they have reported resistance at 31 % via raised MIC.
Further in our study Carbapenemase production was detected in 59.61% of the isolates. This is similar to that reported by Panduragan et al. in 20159 where in has been reported that 62% carbapenemase production amongst isolates.
The highlight of the study was that there were 17.2%(9/52) isolates that had only raised MIC for either of the three carbapenemases namely Ertapenem /Meropenem /Imipnem but did show evidence of carbapenemases production (negative mCIM test).This implies that these bacterial isolates are showing carbapenem resistance by mechanisms other than production of beta lactamases.
Also in these isolates raised MIC for one drug does not necessarily imply resistance to other drugs of the group and so each carbapenem should be indivisually tested before reporting them as resistant or sensitive.
CLSI recommends that unless laboratories can implement the revised carbapenem MIC break points, test for detection of carbapenmases should be performed when isolates of Enterobacteriaceae or Pseudomonas are suspicious of carbapenem production6.
Of all the methods recommended for carbapenemases detection mCIM is the easiest and cheapest to perform and its interpretation is also very objective6,10. The limitation of the method is that it requires overnight incubation as opposed to other methods where results are available within hours.
Through the course of the study we found that 17% of our tested isolates had raised MIC for either of the carbapenems but tested negative for carbapenemase production via mCIM. So we can safely conclude that these 17% isolates had mechanisms other that carbapenemase production as cause of their resistance to carbapenems. It is therefore imperative that all isolates showing raised MIC for carbapenems be tested for production of carbapenemases. However, determination of the mechanism of carbapenem resistance is not advocated as a routine practice for clinical laboratories. However this distinction is important from epidemiological perspective of gram negative isolates and the information is imperative for the successful implementation of any infection control program of hospital.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Doumith M, Ellington MJ, Livermore DM et al. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63(4):659-667.
Crossref - Lartigue MF, Poirel L, Poyart C , et al. Ertapenem resistance of Escherichia coli. Emerg Infect Dis. 2007;13(2):315-317.
Crossref - Martinez-Martinez L. Extended-spectrum b-lactamases and the permeability barrier. Clin Microbiol Infect. 2008;14(suppl 1):82-89.
Crossref - Queenan AM, Bush K. Carbapenemases: the Versatile β-Lactamases. Clin Microbiol Rev. 2007;20(3):440-458.
Crossref - Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect. 2014;44:(2)51-56.
Crossref - Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard – Twelfth Edition. CLSI document M02-A12. Clinical and Laboratory Standards Institute, Wayne, PA. 2019.
- Livermore DM, Woodford N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14(9)413-420.
Crossref - Wattal C, Goel N, Oberoi JK, Raveendran R, Datta S, Prasad KJ. Surveillance of multidrug resistant organisms in a tertiary care hospital in Delhi, India. J Assoc Physicians India. 2010;58:32-36.
- Pandurangan S, BegumEsak S, Narayanasamy A: Phenotypic detection methods of carbapenemase production in Enterobacteriaceae. Int J Curr Microbiol App Sci. 2015;4(6):547-52.
- Mancini S, Kieffer N, Poirel L,Nordmann P. Evaluation of the RAPIDEC(R)® CARBA NP and beta-CARBA(R)® tests for rapid detection of Carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2017;88(4):293-297.
Crossref - Hayajneh WA, Hajj A, Hulliel F, et al. Susceptibility trends and molecular characterization of Gram-negative bacilli associated with urinary tract and intra-abdominal infections in Jordan and Lebanon: SMART 2011-2013. Int J Infect Dis. 2015;35:56-61.
Crossref - Pierce VM,Simner PJ,Lonsway DR,et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among enterobacteriaceae. J Clin Microbiol. 2017; 55: 2321-2333.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.