ISSN: 0973-7510
E-ISSN: 2581-690X
Dengue is one of the most critical diseases, caused by Dengue virus (DENV) serotypes (DENV-1 to DENV-4). Study aims to detect DENV in natural A. aegypti from endemic regions of Uttar Pradesh. From 2010- 2013, mosquitoes collected from Uttar Pradesh tested for Dengue virus serotypes by capsid-premembrane gene-based PCR analyzed for genotypes sequencing of the C-prM junction of DENV genome. A total of 4731, 53.54% (n=2671) A. aegypti and 46 % (n=2060) A. albopictus mosquitoes were collected. Of 226 mosquito pools, 10 pools of A. aegypti and 14 pools of A. albopictus were positive for DENV by PCR. All 24 isolates identified as DENV-I; Genotype (G)-III (n=8), G-V (n=1); DENV-2; G-IV (n=4); DENV-3; G-III (n=11). The overall minimum infection rate was much higher in A. albopictus mosquitoes and presence of MIR in male mosquitoes is an indicating natural vertical transmission and important observation in geographical area indicating natural vertical transmission.
Aedes Mosquitoes, Entomological Surveillance, Dengue Virus, Vertical Transmission, Polymerase Chain Reaction
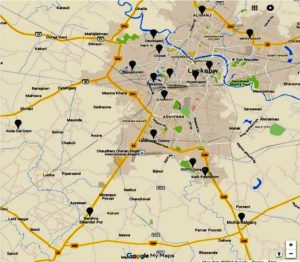
In recent decades, dengue has become a significant threat to human health in tropical and subtropical areas of the world. Aedes aegypti and Aedes albopictus mosquitoes bite humans to deliver the arboviral disease known as DENV.1,2 There are roughly 110 million dengue virus (DENV) infections every year, according to the World Health Organization Report 2023.3 This virus is split into four separate serotypes called DENV-1 to 4 based on its antigenic characteristics. All of the nations in the Indian subcontinent, including India, Pakistan, Bangladesh, and Sri Lanka, have had epidemics brought on by all of the serotypes.4 With a 19.7% dengue positive rate, dengue illness is widespread over much of India, including Uttar Pradesh.5 The study was conducted in the Uttar Pradesh state’s Lucknow region between 2006 and 2010. The sampling was carried out in the geographical areas lies nearby Lucknow (Figure 1). Throughout the five-year trial period, a total of 1227 serum samples were analyzed. 242 of these samples (19.7%) were positive for dengue virus infection between 2006 and 2009. The first outbreak in India to be virologically proven was reported in Calcutta (now Kolkata) between 1963 and 1964.6 Then, in 1996, a sizable and widespread dengue hemorrhagic fever (DHF) pandemic struck the areas surrounding Delhi.7 Since then, the illness has shown a striking return throughout north India, notably Uttar Pradesh. Due to high vector density, the sickness always persisted throughout the year with a surge during monsoon and post-monsoon season.5-7
Significant outbreaks have been recorded from North India in recent years. India has been linked to a change in DENV-2 genotype and a lineage change in DENV-1 genotype.8-11 According to reports from earlier investigations, DENV-3 is the most common strain discovered in the big outbreaks from different locations of India. In contrast to the preceding findings, reports of the resurgence of DENV-4 were also made from Brazil and India.12-14 Increased vector control efforts and a decrease in disease transmission could result from early detection of contaminated mosquitoes.15-16 Identification and typing of DENV from field-caught mosquitoes and larvae are required for epidemiologic research to be successful. Dengue detection in natural Aedes populations has demonstrated to be a viable supplementary tool for early dengue outbreak prediction systems.17-18 Strategies for vector management are essential for preventing or limiting disease transmission because there is no dengue vaccine. As a result, identifying DENV in field-caught mosquito populations is crucial for early warning systems. Given this, the current study’s goal was to identify mosquito viral genetic diversity clusters.
Mosquitoes Collection
Due to the high incidence of DENV and Chikungunya virus (CHIKV) case-patients there,19-20 we collected mosquitoes from a number of places in the Lucknow area where DENV positive patients resided in order to test for DENV in Aedes mosquitoes (Figure 1). From January 2010 to December 2013, adult Aedes mosquitoes were collected in the homes and surrounding regions of DENV-positive patients.21 The pyrethrum spray approach was employed for interior collection, and carbon dioxide-baited traps and battery-operated aspirators were used for outdoor collection. Several collection locations in the Lucknow district were split into urban, suburban, and rural settings. All of the caught adult mosquitoes were brought to the lab, morphologically classified, divided into species and sexes, and kept at -80°C until testing.
Mosquito Identification
Mosquito identification was created in a lab using Rueda’s Aedes species identification keys.22 Microscopic examination led to the identification of the mosquitoes. On a table, mosquitoes were placed in a Petri dish lined with white filter paper. The species were identified using a stereomicroscope using their taxonomic keys, and then counted, sorted, and afterwards separated based on the species, location, and time of collection. Specimens were assigned to a pool based on the species, sex, location, date, and applied collection method.
Mosquito Processing
According to Tome et al., identified mosquitoes were treated between 2010 and 2013.23 Each pool, which included 20–25 mosquitoes or larvae, was homogenized using a motor-driven tissue grinder using 3-5 ml of Media Dulbecco minimum essential medium (MEM), depending on the number of insects. After centrifuging the homogenate, the supernatant was used for further screening.
RNA extraction and RT-PCR
According to Conceicao et al.,24 real-time PCR was used to identify the dengue virus strains. Using the manufacturer’s instructions, RNA was extracted from 140µl of cleared mosquito supernatant using a QI-Aamp viral RNA Mini Kit from Qiagen Hilden, Germany. DENV identification and serotyping using RT-PCR were carried out in mosquito pools as previously mentioned.25
Minimum infection rate (MIR)
The ratio of the number of virus-positive pools to the total number of mosquitoes examined, multiplied by 1000, and was used to calculate the minimum infection rate (MIR).26
Nucleotide sequencing and Phylogenetic analysis
By utilising the QIA-quick Gel Extraction Kit, all PCR positive amplicons were purified (Qiagen, Hilden, Germany). Purified products were cycle sequenced using a Big Dye Terminator cycle sequencing ready reaction kit in accordance with the manufacturer’s instructions (Thermo Fisher Scientific, CA, USA). According to Tripathi et al., 2013,19 the sequencing primers utilized here. The reaction mixture was then fed into an automated DNA sequencer (ABI PRISM 3100 Genetic analyzer, Applied Biosystems, USA) after being purified using a column. The NCBI’s GenBank has received the partial C-prM gene sequences of DENV isolates isolated from mosquito pools. Utilizing the Bio-Edit programme (http://www.mbio.ncsu.edu/bioedit/bioedit.htmL), sequence similarity and identity analysis were conducted. Using the CLUSTAL-W programme (http://www.ebi.ac.uk/clustalw/), the incomplete C-prM gene nucleotide sequences were matched with the sequences of reference strains found in the Gen Bank. Phylogenetic analysis was performed using MEGA 5 software (www.megasoftware.net). The Kimura-two parameter of nucleotide substitution and the neighbor-joining method were used to create the phylogenetic tree. Using bootstrap analysis with 1000 repeats, the tree’s dependability was verified.27
Analysis of environmental factors
Throughout the whole study period, meteorological data including temperature (°C), rainfall (mm), and relative humidity (%) were regularly received from the meteorological department of the Government of India.
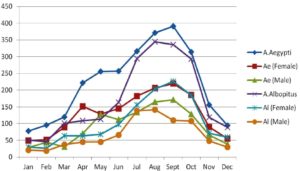
A total of 226 pools of adult Aedes mosquitoes (n=4731) were collected from different sites around the Lucknow district, representing urban and suburban settings (125 pools of A. aegypti and 101 pools of A. albopictus). In comparison to A. albopictus, which had 59 female-only pools and 42 male-only pools out of its 125 total pools, A. aegypti had 72 female-only pools and 53 male-only pools. A. aegypti mosquitoes, numbering 2671 (n=1555 females and n=1116 males), and A. albopictus mosquitoes, numbering 2060 (n=1251 females and n=809 males), respectively (Table 1). Figure 2’s representation of the monthly trend for adult Aedes species reveals that both were most prevalent in the monsoon and post-monsoon seasons, with January, February, and December having the lowest densities.
Figure 2. Monthly trend of Adult mosquito’s indices of A. aegypti and A. albopictus from March 2010- December 2013
Serotype distribution in mosquito pools
Out of a total of 226 positive pools, DENV was detected in 24 pools by RT-PCR (10 pools of adult A. aegypti, n=9 for females; n=1 for males, and 14 pools of adult A. albopictus, n=12 for females; n=2 for males). The serotyping study revealed DENV-1 in 9 pools, DENV-2 in 4 pools, and DENV-3 in 11 pools out of a total of 24 positive pools of Aedes species. However, DENV-1 positive pools were discovered to be at their greatest in 2010, while DENV-2 and DENV-3 positive pools were found to be at their highest in 2012 and 2013. During the time of the study, DENV-4 was not detected. Table 2 displays the DENV serotype distribution by year. In 2010, DENV-1 was the most common serotype; in 2010 and 2012, DENV-2 was more common and in 2013, DENV-1 was replaced by DENV-3. But out of all the serotypes, DENV-3 had the highest incidence in contaminated pools (11/24) (45.83%).
Table (1):
Adult Aedes Mosquitoes collection with their MIR for DENV from May 2010 to October 2013.
| Species | Gender | No. of Mosquitoes | Total No. of Pools | Total No. of Positive Pools by PCR | MIR |
|---|---|---|---|---|---|
| 2010 | |||||
| A. aegypti | Female | 377 | 17 | 4 | 10.61 |
| Male | 290 | 13 | 0 | – | |
| A. albopictus | Female | 314 | 15 | 4 | 12.73 |
| Male | 251 | 13 | 1 | 3.98 | |
| 2011 | |||||
| A. aegypti | Female | 317 | 15 | 1 | 3.15 |
| Male | 261 | 12 | 0 | – | |
| A. albopictus | Female | 301 | 13 | 2 | 6.64 |
| Male | 131 | 7 | 0 | 0 | |
| 2012 | |||||
| A. aegypti | Female | 299 | 14 | 2 | 6.68 |
| Male | 260 | 13 | 0 | – | |
| A. albopictus | Female | 241 | 12 | 2 | 8.29 |
| Male | 174 | 9 | 0 | 0 | |
| 2013 | |||||
| A. aegypti | Female | 562 | 26 | 2 | 3.55 |
| Male | 305 | 15 | 1 | 3.27 | |
| A. albopictus | Female | 395 | 19 | 4 | 10.12 |
| Male | 253 | 13 | 1 | 3.95 | |
| Total | 4731 | 226 | 24 | 5.07 | |
Minimum Infection Rate
Based on the results of PCR performed between 2010 and 2013, the MIR for A. aegypti was estimated to be 3.74 (10/2671 mosquitoes), while the MIR for A. albopictus was 6.79 (14/2060 mosquitoes). Both species’ overall infection rates are predicted to be 5.07 percent. The greatest MIR was determined to be 12.73 (4/314) and 10.12 (4/395) in pools of A. albopictus adult females in 2010 and 2013, respectively, as compared to the pools of A. aegypti (MIR=10.61 & 3.5). MIR of 3.27 has been recorded in A. aegypti, but MIR of 3.98 and 3.95 have been observed in A. albopictus among male mosquitoes in 2010 and 2013, respectively (Table 1).
Table (2):
Year-wise distribution of Dengue serotype in mosquito samples.
Year |
DENV-1 (No. of Pools) |
DENV-2 (No. of Pools) |
DENV-3 (No. of Pools) |
Total No. of Pools |
|---|---|---|---|---|
2010 |
6 |
1 |
2 |
9 |
2011 |
2 |
0 |
1 |
3 |
2012 |
1 |
3 |
0 |
4 |
2013 |
0 |
0 |
8 |
8 |
Total |
9 |
4 |
11 |
24 |
Correlation of environmental factors with DENV incidences
Every year, the latter week of June or the beginning of July saw an increase in dengue fever cases, which peaked from August to October. In Lucknow, the rainy season starts in late June and lasts through October. Regarding changes in maximum temperature (R=-0.17, p=0.959), minimum temperature (R=0.006, p=0.985), and amount of rainfall (R=-0.181, p=0.574), substantial statistical evidence between environmental parameters and DENV IgM positive patients was not found in the current investigation.
Sequence and Phylogenetic analysis
Nucleotide sequencing was used to confirm the DENV serotypes that were isolated from Aedes aegypti and Aedes albopictus. 24 typical dengue viruses’ C-prM gene junction nucleotide sequences from the current investigation were submitted to GenBank, and accession numbers were obtained KM097082 to KM097087, KM097089 to KM097096, KM112168, KM507025 to KM507026, and KM507029-KM507035 are the samples’ GenBank accession numbers. In this investigation, 8 of the 9 DENV-1 strains were genotype III, and one was genotype V. While all 11 DENV-3 strains were classified as genotype III, all 4 DENV-2 strains belonged to genotype IV. Table 3 provides a summary of the genotyping data results along with their accession number.
Table (3):
Accession Number and Dengue serotype/genotype in collected mosquitoes.
S. No. |
Accession Number |
Year |
Vector species |
Serotype |
Genotype |
|---|---|---|---|---|---|
1 |
KM097082 |
2010 |
A. aegypti |
DENV-1 |
III |
2 |
KM097083 |
2010 |
A. aegypti |
DENV-1 |
III |
3 |
KM097084 |
2010 |
A. albopictus |
DENV-1 |
III |
4 |
KM097085 |
2010 |
A. albopictus |
DENV-1 |
III |
5 |
KM097086 |
2010 |
A. albopictus |
DENV-1 |
III |
6 |
KM097087 |
2010 |
A. albopictus |
DENV-1 |
III |
7 |
KM507025 |
2010 |
A. aegypti |
DENV -3 |
III |
8 |
KM507026 |
2010 |
A. albopictus |
DENV -2 |
IV |
9 |
KM507034 |
2010 |
A. aegypti |
DENV -3 |
III |
10 |
KM097089 |
2011 |
A. aegypti |
DENV-1 |
V |
11 |
KM097090 |
2011 |
A. albopictus |
DENV-1 |
III |
12 |
KM507035 |
2011 |
A. albopictus |
DENV -3 |
III |
13 |
KM097091 |
2012 |
A. aegypti |
DENV-1 |
III |
14 |
KM112168 |
2012 |
A. albopictus |
DENV-2 |
IV |
15 |
KM507029 |
2012 |
A. aegypti |
DENV-2 |
IV |
16 |
KM507030 |
2012 |
A. albopictus |
DENV-2 |
IV |
17 |
KM097092 |
2013 |
A. aegypti |
DENV-3 |
III |
18 |
KM097093 |
2013 |
A. aegypti |
DENV-3 |
III |
19 |
KM097094 |
2013 |
A. albopictus |
DENV-3 |
III |
20 |
KM097095 |
2013 |
A. albopictus |
DENV-3 |
III |
21 |
KM097096 |
2013 |
A. albopictus |
DENV-3 |
III |
22 |
KM507031 |
2013 |
A. albopictus |
DENV-3 |
III |
23 |
KM507032 |
2013 |
A. albopictus |
DENV-3 |
III |
24 |
KM507033 |
2013 |
A. aegypti |
DENV-3 |
III |
DENV-1
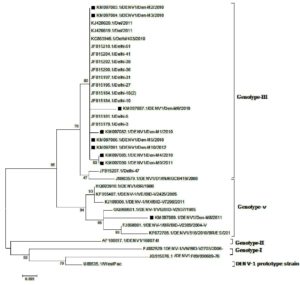
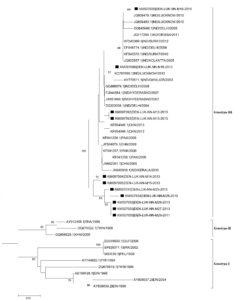
A phylogenetic tree based on partial C-prM gene sequences was created using 24 reference DENV-1 strains that were acquired from Gen Bank and 9 mosquito-derived DENV-1 sequences. This tree showed that isolates from India and other countries fell into two separate genotypes (III and V) (Figure 3). According to phylogenetic research, 8 of the 9 DENV-1 strains (KM097082–KM097087, KM097090, KM097091) isolated from mosquitoes shared a 99.4%–99.7% nucleotide sequence similarity with the human DENV-1 strain from Delhi 2010 and are all genotype III strains. One DENV-1 strain (KM97089) from a mosquito is aggregating with strains of Genotype V at the same time.
Figure 3. Phylogenetic tree of studied Dengue serotype 1 sequences was constructed by Maximum Likelihood method (1,000 Bootstrap replicates) in MEGA 5.05 software. The tree is based on C–prM regions of the selected strains. The studied strains of DENV-1 serotype are indicated by black square shaped box prior to the accession number of the studied
DENV-2
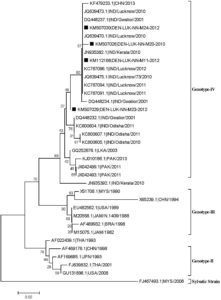
A Phylogenetic tree based on incomplete C-prM gene sequences was created using 31 reference strains of DENV-2 that were obtained from Gen Bank, and it revealed that isolates from India and other countries fell into three separate genotypes (II, III, and IV) (Figure 4). Based on phylogenetic analysis, we discovered that the four DENV-2 strains (KM50726, KM507029, KM507030, and KM112168) isolated from mosquitoes shared 99.4%–99.7% of their amino acid sequences with one another and 99.2%–99.9% of their amino acid sequences with the genotype IV human DENV-2 strains from Lucknow, Kerala, Gwalior, and Lucknow 2010–2012.
Figure 4. Phylogenetic tree of studied Dengue serotype 2 sequences was constructed by Maximum Likelihood method (1,000 Bootstrap replicates) in MEGA 5.05 software. The tree is based on C–prM regions of the selected strains. The studied strains of DENV-2 serotype are indicated by black square shaped box prior to the accession number of the studied sequences. Bootstrap values of >40 are shown in the tree. Scale bar indicates the nucleotide substitutions per site
DENV-3
A Phylogenetic tree based on partial C-prM gene sequences was created using 33 reference strains of DENV-3 that were obtained from Gen Bank, and 11 mosquitoes-derived DENV-3 sequences. This revealed that isolates from India and other countries around the world clustered into three different genotypes (I, II, and III) (Figure 5). All of the DENV-3 (n=11) strains used in this investigation had a 99.2 to 99.9% nucleotide similarity. According to a phylogenetic analysis of the C- prM gene sequence, all 11 DENV-3 mosquito-derived genotype-III strains, as well as strains from India, China, and Pakistan, fall into two sub-clusters (KM507025, KM507031-KM507035, and KM097092-KM097096).
Figure 5. Phylogenetic tree of studied Dengue serotype 3 sequences was constructed by Maximum Likelihood method (1,000 Bootstrap replicates) in MEGA 5.05 software. The tree is based on C–prM regions of the selected strains. The studied strains of DENV-3 serotype are indicated by black square shaped box prior to the accession number of the studied sequences. Bootstrap values of >40 are shown in the tree. Scale bar indicates the nucleotide substitutions per site
Over the past 20 years, dengue, an acute arboviral illness, has grown to be a significant public health threat in tropical and subtropical areas. Mosquito species Aedes aegypti and Aedes albopictus are the main urban vectors involved in the transmission of the dengue virus around the world, including India. Vector control measures are essential for avoiding or limiting disease transmission given the paucity of dengue vaccinations. Therefore, it has become clear that isolating and identifying DENV from field-caught mosquitoes is a crucial component of an early warning system. In the current work, we examined DENV infection in Aedes vectors and their genetic characteristics in the Lucknow District around DENV affected areas from March 2010 to October 2013. Several papers had previously described the DENV activity in this area,28-29 referring to the region’s predominance of mosquito vectors as the reason for DENV persistence in the Lucknow district. Entomological observation of adult Aedes mosquitoes in the Lucknow district throughout our investigation revealed that 43.54% of them belonged to the Aedes albopictus population and 56.45% to the Aedes aegypti population. In adult female Aedes aegypti and Aedes albopictus mosquitoes, DENV-1 to 3 was isolated and genotypically identified for the first time in Uttar Pradesh.
According to our investigation, A. aegypti was most prevalent in the studied area,30 which is consistent with the findings of the other studies. During the dry season, both species were confined to a small number of nesting locations. Even though, compared to A. aegypti, the larval species of A. albopictus were pronouncedly growing during and after the monsoon.24 positive pools were DENV positive out of the total of 4731 mosquitoes, proving DENV transmission in this area. While DENV-2 circulated at a lesser frequency, DENV-1 and DENV-3 were the two most common serotypes in 2010 and 2013, respectively.31 In 2010, the mosquitoes that had been captured tested positive for all three serotypes (DENV-1, 2 and 3). In the same year, similar serotypes were discovered in human samples; 60% of dengue cases were DENV-1, 1.8% was DENV-2, and 24.3% were DENV-3,32-33 According to studies, adult Aedes aegypti and Aedes albopictus mosquitoes from Orissa and Surat between 2010 and 2011 had DENV-2 and DENV-3.34-35 DENV-3 (n=8) was found to be the most common serotype in 2013. According to our research, DENV-1 predominated in 2010–2011, followed by DENV 2 in 2012, and DENV 3 in 2013. DENV-1 and 2 were then replaced by DENV-3 in 2014.
Therefore, the current study demonstrates that during many year-wise outbreaks, the prevalence of each serotype within a certain geographic area has altered. The three serotypes are circulating with the shift in dominant species, according to results from other studies.10,36 To define the molecular subtype/genotype of this virus and the introduction of novel genotypes, molecular characterization is required. The C-prM gene junction has been described as a potent genotyping tool in many studies.37-39 Eight of the nine DENV-1 derived strains sequenced in this work were assigned to genotype III by phylogenetic analysis, placing them in close proximity to previous Indian human DENV-1 infections reported from the Delhi and Lucknow region in 2010. There are many of DENV-1 genotype III viruses in Mexico, Colombia, Brazil, and Venezuela. Because the viruses recovered in 2001–2002, 2004–2005, and 2006–2007 from northern India belong to different lineages, the majority of Indian DENV-1 genotype III viruses are phylogenetically distinct. Additionally, one Lucknow strain (KM097089) generated from a mosquito and discovered in 2011 corresponded to genotype-V. This genotype is typically found in Asian nations, African and American countries.40,41 The genotype IV status of the four DENV-2 mosquito-derived strains from 2010–2012 allowed them to be more closely related to earlier isolates from Lucknow (2010), Kerala (2010), and Gwalior (2011) than to any other isolates.
In India, genotype IV was initially discovered in New Delhi11-17 during the dengue epidemic of 1996. Human isolates from Gwalior in 2001, Delhi in 2007–2009, and Kerala in 2008–201042 have already been connected to DENV-2 genotype IV. Our findings show that DENV-2 genotype IV is the most common genotype derived strain in this area. Each of the eleven DENV-3 strains sequenced in this study was given the genotype III. The Indian strains that have previously been described from Chennai (2013), Orissa (2011), and Surat (2012)42-43 are closely related to these eleven mosquito-derived strains. The most prevalent DENV-3 genotype44 circulating worldwide is genotype III. This study demonstrates that genotype III is the most prevalent strain of DENV-3 coming from a mosquito in this region. A. aegypti and A. albopictus were found to be the two most common species, according to entomological research conducted in the areas under study. MIR in A. aegypti and A. albopictus was evaluated from 2010 to 2013. We discovered a 12.73 fold rise in MIR in A. albopictus in 2010, followed by a 10.12 fold increase in 2013.16 This is similar to recent results from India. MIR varies depending on a number of variables, including the collecting period, the size of the sample pool, and the quantity of processed samples.34
Our study’s key conclusion is the identification of DENV in mosquitoes, particularly male mosquitoes. Since male mosquitoes do not feed on blood, the vertical (trans-stadial or transovarial) transmission phenomena32-40 is the most likely method by which male mosquitoes can become infected. Humans are exposed to the virus by venereal transmission, which occurs when an adult male mosquito transmits it to a female A. aegypti during mating.45 Vertical transmission in conjunction with venereal transmission is therefore likely to act as a survival mechanism to keep this virus in the environment at the research locations in the Lucknow district. Research evidence regarding arbovirus transmission from women to men has grown. Critical factors for arbovirus vertical transmission in mosquitoes were studied by Lequime and Lambrechts in 2014 and by Leguime et al.46-47 An explanation of the vertical transmission of arbovirus in mosquitoes is given by Mao et al. In this part of India,48 there have never been any reports of DENV being found in Aedes mosquito populations. As a result of the co-circulation of several DENV serotypes in Northern India, this study demonstrates the growing endemicity of the disease. The current study provides the first evidence of DENV in field-caught mosquitoes in this area. A well-functioning early warning system for dengue outbreaks is necessary to control these vectors in this area, as high MIR found in male Aegypti and A. albopictus mosquitoes is indicative of natural vertical transmission.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- World Health Organization; 2009. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. World Health Organization. https://apps.who.int/iris/handle/10665/44188
- Smith KM., Nanda K, Spears CJ, et al. Structural mutants of dengue virus 2 transmembrane domains exhibit host-range phenotype. Virol J. 2011;9(8):289.
Crossref - World Health Organization; 2023, Dengue and severe dengue. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue
- Raheel U, Faheem M, Riaz MN, et al. Dengue fever in the Indian Subcontinent: an overview. J Infect Dev Ctries. 2011;5(4):239-247.
Crossref - Garg A, Garg J, Rao YK. et al. Prevalence of dengue among clinically suspected febrile episodes at a teaching hospital in North India. Journal of Infectious Diseases and Immunity. 2011;3(5):85-89.
- Ramakrishnan SP, Gelfand HM, Bose PN, Sehgan PN, Mukherjee RN. The Epidemic of Acute Haemorrhagic Fever, Calcutta, 1963: Epidemiological Inquiry. Indian J Med Res. 1964;52:633-650.
- Dar L, Broor S, Sengupta S, et al. The first major outbreak of dengue hemorrhagic fever in Delhi, India. Emerg Infect Dis. 1999;5(4):589-590.
Crossref - Pandey N, Nagar R, Gupta SO, et al. Trend of dengue virus infection at Lucknow, north India (2008- 2010): a hospital based study. Indian J Med Res. 2012;136(5):862-867.
- Dash PK, Parida MM, Saxena P, et al. Re-emergence of dengue virus type-3 (subtype-III) in India: implications for increased incidence of DHF & DSS. Virol J. 2006;3:55.
Crossref - Kukreti H, Dash PK, Parida M, et al. Phylogenetic studies reveal existence of multiple lineages of a single genotype of DENV-1 (genotype III) in India during 1956-2007. Virol J. 2009;6(6):1.
Crossref - Kumar R, Jyotsana C, Garima A, et al. Changing clinical manifestations of dengue infection in north India. WHO Regional Office for South-East Asia, 2008.
- Patil JA, Cherian S, Walimbe AM, et al. Evolutionary dynamics of the American African genotype of dengue type 1 virus in India (1962-2005). Infect Genet Evol. 2011;11(6):1443-1448.
Crossref - de Melo FL, Romano CM, de Andrade Zanotto PM. Introduction of dengue virus 4 (DENV-4) genotype I into Brazil from Asia? PLoS Negl Trop Dis. 2009;3 (4):e390.
Crossref - Cecilia D, Kakade MB, Bhagat AB, et al. Detection of dengue-4 virus in pune, western india after an absence of 30 years–its association with two severe cases. Virol J. 2011;8:46.
Crossref - Das B, Sahu A, Das M, et al. Molecular investigations of chikungunya virus during outbreaks in Orissa, Eastern India in 2010. Infect Genet Evol. 2010;12(5):1094-101.
Crossref - Chow VT, Chan YC, Yong R, et al. Monitoring of dengue viruses in field-caught Aedes aegypti and Aedes albopictus mosquitoes by a type-specific polymerase chain reaction and cycle sequencing. Am J Trop Med Hyg. 1998;58(5):578-586.
Crossref - Fouque F, Garinci R, Gaborit P, Epidemiological and entomological surveillance of the co-circulation of DEN-1, DEN-2 and DEN-4 viruses in French Guiana. Trop Med Int Health. 2004;9(1):41-46.
Crossref - Urdaneta L, Herrera F, Pernalete M, et al. Detection of dengue viruses in field-caught Aedes aegypti (Diptera: Culicidae) in Maracay, Aragua state, Venezuela by type-specific polymerase chain reaction. Infect Genet Evol. 2005;5(2):177-184.
Crossref - Tripathi SK, Gupta P, Khare V, et al. Emergence of new lineage of Dengue virus 3 (genotype III) in Lucknow, India. Iran J Microbiol. 2013;5(1):68-75.
- Nyari N, Maan HS, Sharma S, Pandey SN, Dhole TN. Identification and genetic characterization of chikungunya virus from Aedes mosquito vector collected in the Lucknow district, North India. Acta Trop. 2016;158:117-124.
Crossref - Ghavami MB. Estimation and Comparison of Anopheles maculipennis s.l. (Diptera: Culicidae) Survival Rates with Light-trap and Indoor Resting Data. Iran J Public Health. 2005;34(2):48-57.
- Reuben R, Tewari SC, Hiriyan J, Akiyama J. Illustrated keys to species of Culex (Culex) associated with Japanese encephalitis in Southeast Asia (Diptera: Culicidae). Mosquito Systematic. 1994;26(2):75-96.
- Tome HV, Pascini TV, Dangelo RA, Guedes RN, Martins GF. Survival and swimming behavior of insecticide-exposed larvae and pupae of the yellow fever mosquito Aedes aegypti. Parasit Vectors. 2014;27:195.
Crossref - Conceicao TM, Da Poian AT, Sorgine MH. A real-time PCR procedure for detection of dengue virus serotypes 1, 2, and 3, and their quantitation in clinical and laboratory samples. J Virol Methods. 2010;163(1):1-9.
Crossref - Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30(3):545-551.
Crossref - Savage HM, Smith GC, Moore CG, Mitchell CJ, Townsend M, Marfin AA. Entomologic investigations of an epidemic of St. Louis encephalitis in Pine Bluff, Arkansas, 1991. Am J Trop Med Hyg. 1993;49(1):38-45.
Crossref - Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-2379.
Crossref - Mishra G, Jain A, Prakash O, et al. Molecular characterization of dengue viruses circulating during 2009-2012 in Uttar Pradesh, India. J Med Virol. 2015;7(1):68-75.
Crossref - Ansari MA, Razdan RR. Seasonal prevalence of Aedes aegypti in five localities of Delhi, India. Dengue Bull, 2020;22:28-32.
- Almeida AP, Baptista SS, Sousa CA, et al. Bioecology and vectorial capacity of Aedes albopictus (Diptera: Culicidae) in Macao, China, in relation to dengue virus transmission. J Med Entomol. 2005;42(3):419-428.
Crossref - Afreen N, Naqvi IH, Broor S, Ahmed A, Parveen S. Phylogenetic and Molecular Clock Analysis of Dengue F19type 1 and 3 from New Delhi, India. PLoS One. 2015;10(11):e0141628.
Crossref - Ahmed NH, Broor S. Dengue Fever outbreak in delhi, north India: a clinico-epidemiological study. Indian J Community Med. 2015;40(2):135-138.
Crossref - Paingankar MS, Gokhale MD, Vaishnav KG, Shah PS, Monitoring of dengue and chikungunya viruses in field-caught Aedes aegypti (Diptera: Culicidae) in Surat city, India. Current Science. 2014;10:1559-1567.
- Agarwal R, Kapoor S, Nagar R, et al. A clinical study of the patients with dengue hemorrhagic fever during the epidemic of 1996 at Lucknow, India. Southeast Asian J Trop Med Public Health. 1999;30(4):735-740.
- Anoop M, Issac A, Mathew T, et al. Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South India. Indian J Exp Biol. 2010;48(8):849-857.
- Aviles G, Rowe J, Meissner JM, Caffarena JCM, Enria D, Jeor SS. Phylogenetic relationships of dengue-1 viruses from Argentina and Paraguay. Arch Virol. 2002;47(11):2075-2087.
Crossref - Descloux E, Cao-Lormeau VM, Roche C, Hamballerie XD. Dengue 1 diversity and microevolution, French Polynesia 2001-2006: connection with epidemiology and clinics. PLoS Negl Trop Dis. 2009;3(8):e493.
Crossref - Fu J, Tan BH, Yap EH, Chan YC, Tan YH. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90). Virology. 1992;188(2):953-958.
Crossref - Singh UB, Maitra A, Broor S, Rai A, Pasha ST, Seth P. Partial nucleotide sequencing and molecular evolution of epidemic causing Dengue 2 strains. J Infect Dis. 1999;180(4):959-965.
Crossref - Singh UB, Seth P. Use of nucleotide sequencing of the genomic cDNA fragments of the capsid/premembrane junction region for molecular epidemiology of dengue type 2 viruses. Southeast Asian J Trop Med Public Health. 2001;32(2):326-335.
- Dash PK, Sharma S, Srivastava A, et al. Emergence of dengue virus type 4 (genotype I) in India. Epidemiol Infect. 2011;139(6):857-861.
Crossref - Guedes DR, Cordeiro MT, Melo-Santos MA, et al. Patient-based dengue virus surveillance in Aedes aegypti from Recife, Brazil. J Vector Borne Dis. 2010;47(2):67-75.
- Thavara U, Siriyasatien P, Tawatsin A, Double infection of hetero-serotypes of dengue viruses in field populations of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and serological features of dengue viruses found in patients in southern Thailand. Southeast Asian J Trop Med Public Health. 2006;37(3):468-476.
- Yergolkar PN, Cherian SS Jadhav S, Rant CG, Mourya DT. Genetic characterization of dengue virus types 1 and 2 in India, with emphasis on the viruses circulating in Karnataka. Indian J Med Res. 2017;146(5):662-665.
Crossref - Mavale M, Parashar D, Sudeep A, et al. Venereal transmission of chikungunya virus by Aedes aegypti mosquitoes (Diptera: Culicidae). Am J Trop Med Hyg. 2010;83(6):1242-1244.
Crossref - Lequime S, Lambrechts L. Vertical transmission of arboviruses in mosquitoes: a historical perspective. Infect Genet Evol. 2014;28:681-690.
Crossref - Lequime S, Paul RE, Lambrechts L. Determinants of Arbovirus Vertical Transmission in Mosquitoes. PLoS Pathog. 2016;12(5):e1005548.
Crossref - Mao Q, Wu W, Liao Z. et al. Viral pathogens hitchhike with insect sperm for paternal transmission. Nat Commun. 2019;10(1):955.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.